Initial report on the search for Steamer retroelement sequence in Global Ocean Sampling (GOS) data. Inspired by This Week in Virology episode 337: "a transmissible tumor in soft shell clams associated with a retrovirus-like element in the clam genome."
If you're impatient you can skip to the end.
From the PNAS article we obtain the accession number KF319019. We download the sequence Mya arenaria retrotransposon steamer GagPol gene, complete cds in FASTA format. We also download the translated GagPol protein sequence, accession AIE48224.1 in FASTA format.
Before embarking on further computational work, we try BLAST of the query sequences at the NCBI website.
Neither nucleotide vs. nucleotide (blastn, nr database) nor protein vs. translated nucleotide (tblastn, nr database ocean metagenome subset) returned results. However, protein vs. protein (blastp, metagenomic proteins env_nr database) returned results with hits to GOS indicating that further work is worthwhile.
Data from the Global Ocean Sampling project are available at the iPlant datastore. We downloaded the metadata in CSV format and the assembled contigs (17 GB) in FASTA format.
On attempting to create a local BLAST database from the GOS contigs, we found that the file contained many duplicate sequences. A new non-redundant FASTA file was generated using the Bioperl utility bp_nrdb:
bp_nrdb -o gosAsmNR.fa CAM_PROJ_GOS.asm.fa
Now we can create the BLAST database (version 2.2.30+):
makeblastdb -in gosAsmNR.fa -dbtype nucl -parse_seqids -hash_index -title GOSasmNR -out GOSasmNR
And run blastn, tblastn and tblastx, with tab-separated output:
blastn -db GOSasmNR -query data/KF319019.1.fa -out data/steamer_v_gosasm_blastn.tsv -outfmt 6 -num_threads 4
tblastn -db GOSasmNR -query data/AIE48224.1.fa -out data/steamer_v_gosasm_tblastn.tsv -outfmt 6 -num_threads 4
tblastx -db GOSasmNR -query data/KF319019.1.fa -out data/steamer_v_gosasm_tblastx.tsv -outfmt 6 -num_threads 4
The results, with number of hits where E <= 10 (the default value):
The default blastn algorithm is megablast (expects high similarity) so 0 hits is not surprising. Using -task blastn returns hits but the best E = 0.003 across only short (~ 80 nt) alignment lengths.
First we extract column 2 (hits) from the BLAST output files and write the (unique) accessions to new files:
cut -f2 data/steamer_v_gosasm_tblastn.tsv | uniq > data/tblastn_hits.txt
cut -f2 data/steamer_v_gosasm_tblastx.tsv | uniq > data/tblastx_hits.txt
Next we use these files to dump the header line of the corresponding sequence entries from the BLAST database to new files:
blastdbcmd -db GOSasmNR -dbtype nucl -entry_batch data/tblastn_hits.txt | grep "^>" > data/tblastn_headers.txt
blastdbcmd -db GOSasmNR -dbtype nucl -entry_batch data/tblastx_hits.txt | grep "^>" > data/tblastx_headers.txt
Now we have all the files we need in the data directory for the final step.
The header lines for (some of) the GOS sequences contain both the sequence accession and a sample ID in this form:
/sample_id=JCVI_SMPL_1103283000001
(Some of) these sample IDs are also present in the GOS metadata file, which contains sample latitude and longitude. So all we need to do is match the accessions from the BLAST output with those in the sequence headers, then match the sample IDs from the headers with those in the metadata file, and so link BLAST hit accessions with geographic coordinates.
We do this using the R code in steamer.R.
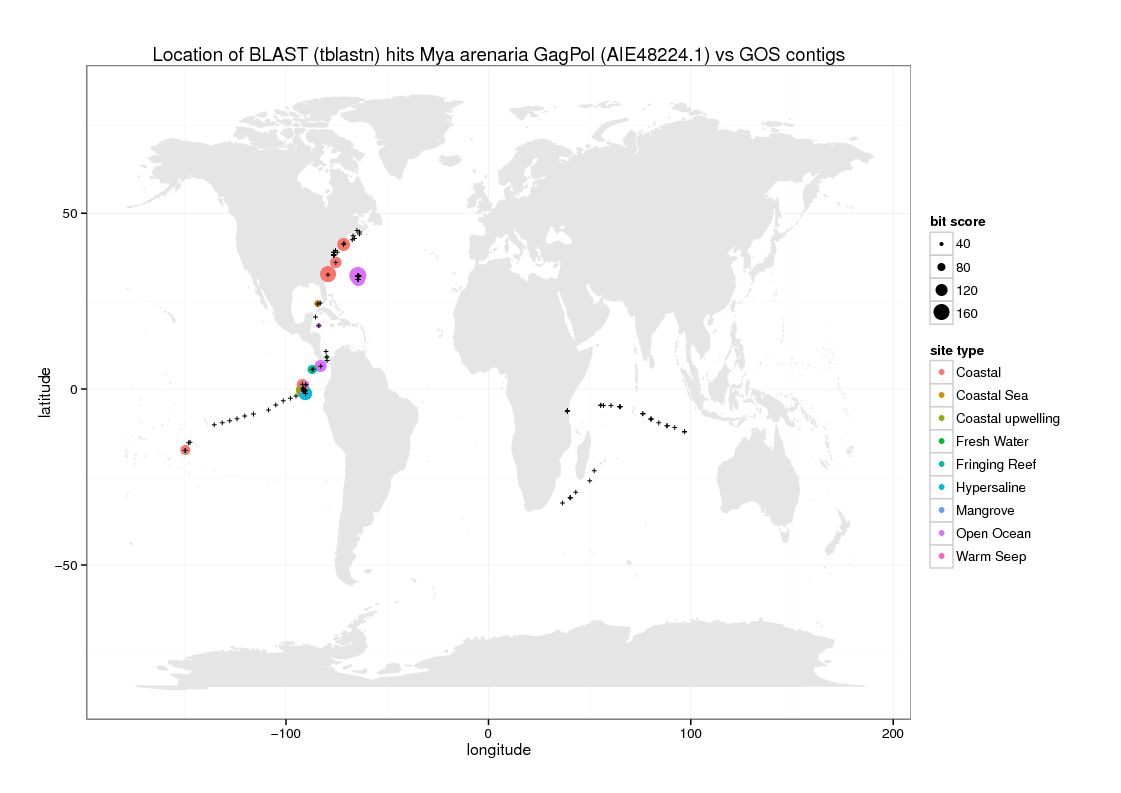
There are many ways to plot maps using R. I just used a solution obtained from a quick Google search, to plot those hits from the tblastn output file with matching coordinates, using size to indicate bit score and color to indicate the site description. Note that only 192/347 tblastn hits have this information.
Here are the results: PDF file | PNG file
- Note that the BLAST hits are not especially convincing: they are low identity with gaps and only protein vs translated nucleotide returns results, so it is not clear whether they represent something similar to Steamer GagPol or something else
- This initial visualization needs some work to improve the clarity, but goes some way to illustrating the position of the best BLAST hits; a more dynamic visualization using e.g. Google Maps would be an improvement
- There is some indication that the best hits are located mostly close to North/Central America and are often coastal, but it is difficult to conclude the significance as the data are quite limited