New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[REVIEW]: GAMA: Genetic Automated Machine learning Assistant #1132
Comments
|
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @jsgalan it looks like you're currently assigned as the reviewer for this paper 🎉. ⭐ Important ⭐ If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿 To fix this do the following two things:
For a list of things I can do to help you, just type: |
|
|
Just making sure I did not miss anything, but there is not something I should be doing right now, correct? |
Correct, we're waiting on @jsgalan to complete their review (by updating the checklist above). |
|
@jsgalan |
|
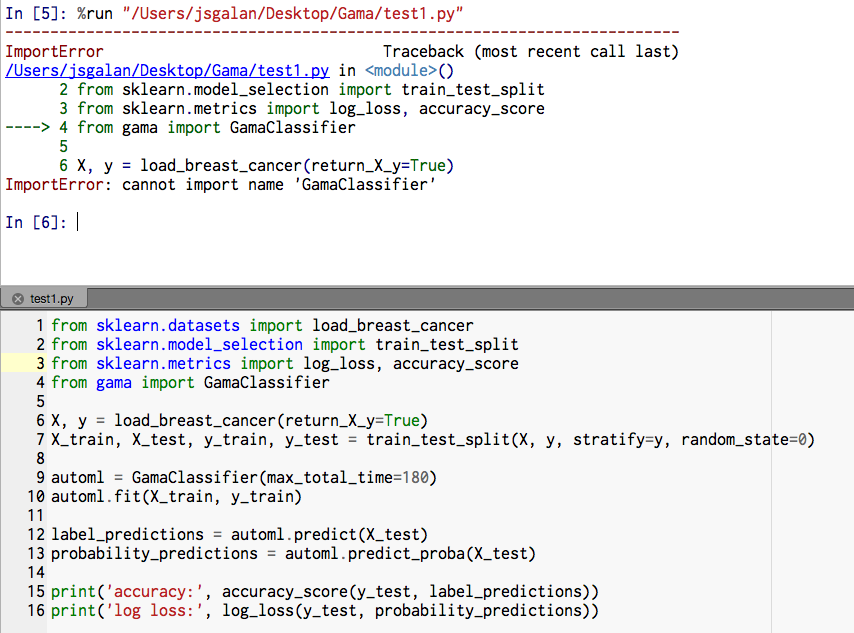
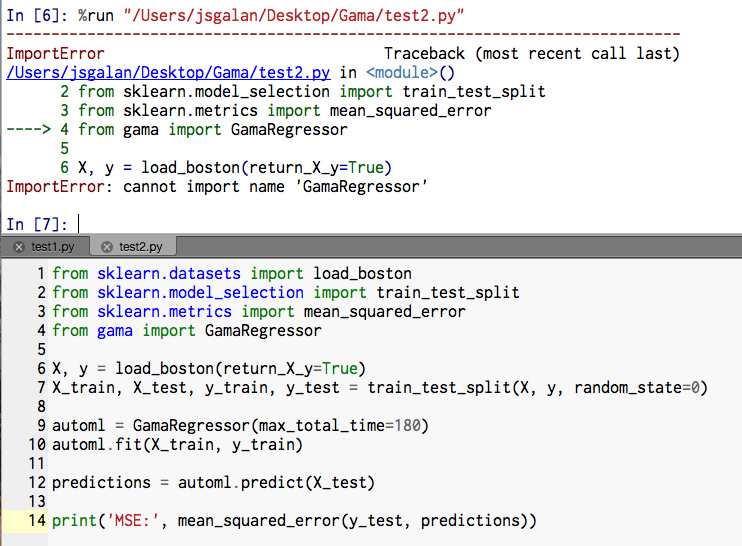
Hi all, i proceeded with the first steps of the review (installation and upgrades here: Everything is installed correctly, however I get the following errors, Any clues? Best |
|
That's odd, I can not recreate the issue locally (tried a clean install on a ubuntu docker image). It looks like you installed it to a 'gama' virtual environment, are you also running the scripts from that environment? |
|
Hi, i restarted the computer and everything worked fine :) Here are my results for the tests provided in the documentation: Had a few warnings for run-test2.txt and run-test5.txt And the rest looked fine also for |
|
Good to hear it works now! Thank you for the feedback, I will ping you here when I am done resolving the issues. I expect to be able to finish them all by tomorrow. |
|
Hi @jsgalan!
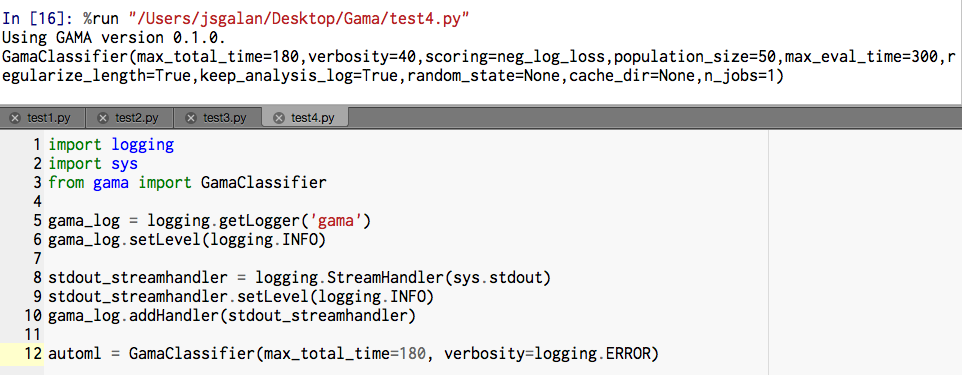
I changed log levels of some statements so that anything of at least Thanks again for your time. |
|
Hi all, Sorry for being so vague on my descriptions. I will reformulate: 1- add a few examples (not just the minimal example, but all five examples) to the Github repository. 1.1-Also please check that the examples is wrongly edited, missing the last part of the statement <-- this was done! [check!] 2- What more detailed explanation are you missing for the log visualization? <- can this link or this information be found somewhere on the initial Github webpage 3- We mentioned the log visualization and type of questions you can currently answer with it in our paper submission. What in particular were you missing or expected to be changed? The article currently reads:
I think is important to include in those lines regarding the aspects such as: a) the figures/plots it produces. (a.e Fitness over number of evaluations vs No. evaluations, Pipeline length over number of evaluations vs No. evaluations and more importantly how Fitness over number of evaluations by main learner vs No. evaluations ) b) all the methods/models that can be learned (a.e NB, Decision Tree,Boosting, Random Forest, KNN, SVC, LogReg). This is not stated clearly in the documentation. Sorry for being stubborn but I think this should be main focus of the article and be very well described in the Github website, showing all the full capabilities of the software and all the models that can be tested using the implementation. After revising I think some extra homework... 4- There were .csv files generated that are not well described anywhere 4.1- There were folders empty folders created with the same name of the .csv files. Hope this helps. |
Could you elaborate what you mean specifically? I made three of them into separate scripts in the example folder and linked them from the
It is referred to in the documentation but I will add it to the
I turned it off for examples now, as I don't think examples should produce extra files. I will add documentation regarding the files should you wish to produce them.
Yes, using the 'cache_dir' hyperparameter when initializing a I'll make the changes tomorrow. |
Also, no worries :) it's your job 👍 I appreciate the effort you put in |
|
@whedon generate pdf |
|
|
@whedon generate pdf |
|
|
@whedon generate pdf Sorry for the spam. It seems that Whedon did not take the last version last time? Missing a line-break. |
|
|
Hi @jsgalan,
No news since last comment.
Added link and example image to the
I added a plot example to the paper and also explicitly named some algorithms as well as emphasize using scikit-learn algorithms (see the latest article proof).
I find the adjustments I made are a reasonable balance between providing too little information and all information. Please let me know if you disagree.
They now have their section in the documentation. |
|
Hi all @arfon @PGijsbers I just revised and all the changes suggested were made in documents and pdf, respectively. I am happy to say that from my part all the requirements were fulfilled.
Answering to this comment, after my initial thoughts and replies, I imagined that the package can be (and will be extended) to include different types of tests or can be put into a greater pipeline using various packages, so i concur with your reasoning. Happy to revise and test this novel tool that will be beneficial for the ML community. Best |
|
Thanks again for all your effort @jsgalan! |
|
Seems like it is all fixed @arokem |
|
@whedon set 10.5281/zenodo.2545472 as archive |
|
I'm sorry @PGijsbers, I'm afraid I can't do that. That's something only editors are allowed to do. |
|
Actually not sure what is preferred here:
|
|
I think that this one is fine: 10.5281/zenodo.2545472 This is the version you generated after incorporating all of the reviewer comments? Could you please edit the metadata of the Zenodo page (title and authors), so that it matches the paper? Thanks! |
|
Done! |
|
@whedon set 10.5281/zenodo.2545472 as archive |
|
OK. 10.5281/zenodo.2545472 is the archive. |
|
Congratulations! Your paper is now ready to be accepted. Stand by for EIC or an Associate EIC to drop by and finalize this. |
|
Thanks for editing @arokem |
|
@whedon accept |
|
|
|
Check final proof 👉 openjournals/joss-papers#460 If the paper PDF and Crossref deposit XML look good in openjournals/joss-papers#460, then you can now move forward with accepting the submission by compiling again with the flag |
|
@whedon accept deposit=true |
|
|
🚨🚨🚨 THIS IS NOT A DRILL, YOU HAVE JUST ACCEPTED A PAPER INTO JOSS! 🚨🚨🚨 Here's what you must now do:
Any issues? notify your editorial technical team... |
|
🎉🎉🎉 Congratulations on your paper acceptance! 🎉🎉🎉 If you would like to include a link to your paper from your README use the following code snippets: This is how it will look in your documentation: We need your help! Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
|







Submitting author: @PGijsbers (Pieter Gijsbers)
Repository: https://github.com/PGijsbers/GAMA
Version: v0.1.0
Editor: @arokem
Reviewer: @jsgalan
Archive: 10.5281/zenodo.2545472
Status
Status badge code:
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@jsgalan, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
The reviewer guidelines are available here: https://joss.theoj.org/about#reviewer_guidelines. Any questions/concerns please let @arokem know.
✨ Please try and complete your review in the next two weeks ✨
Review checklist for @jsgalan
Conflict of interest
Code of Conduct
General checks
Functionality
Documentation
Software paper
paper.mdfile include a list of authors with their affiliations?The text was updated successfully, but these errors were encountered: