New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Highlighting features used by prediction path #75
Comments
|
Hi @Rkubinski, thanks for your feedback. |
|
Great ! Thanks :) |
|
Weird. Is that on windows? mac? Does it do same if vertical? |
|
Same problem with vertical, this is on Ubuntu 18.10, |
|
Ok, this was a font metrics problem when it was happening on mac/windows. Maybe we need to make that configurable. or if already is, please try diff font. |
|
Where do I pass a font type option ? Dont see such a parameter for dtreeviz object |
|
@Rkubinski can you provide the list of feature names ? I tried to reproduce the issue with long feature names and it works. |
dtreeviz() method has the 'fontname' parameter |
|
Ok, I was not sure what you meant by font metrics. OTUs: |
|
@Rkubinski, indeed you have a long list of features :). Meanwhile I will try to reproduce the issue with a list of features similar in length with yours ;) |
|
I created a vertical version for instance table. It is configurable through instance_orientation parameter. @parrt do you have a better name for instance_orientation parameter ? |
|
Hi, |
|
I plan to make a PR tomorrow :) |
|
|
|
@Rkubinski I have just created the PR #78 . I hope it will solve your issue. Waiting for your feedback ;) |
|
Hi guys. Thank you so much for the help. I am still new to this, I tried to update dtreeviz via pip but I guess that has not been updated yet. So to use this new version, I am assuming I would have to pull from github and run the scripts directly ? |
|
Hi @Rkubinski, yes, you can use the master version from repo until @parrt will make a new release. |
|
@tlapusan Great! Ill get back to you when Ive tested it out |
|
hi @Rkubinski, any feedback ? I would like to close this issue. |
|
Hi guys, sorry for the delay in feedback, it did work out in the end :) |
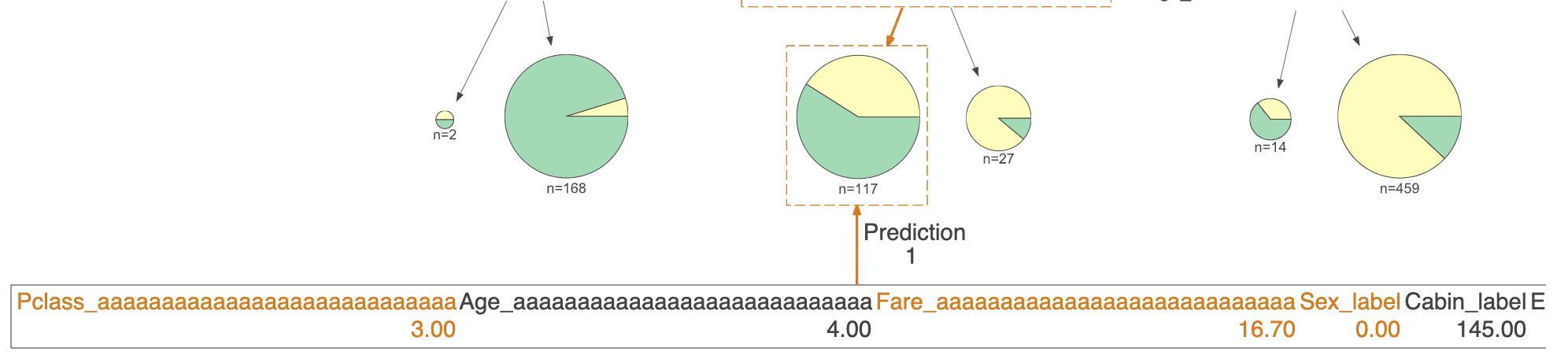

Is there any way to remove highlighted features used by prediction path or print them differently like in a table along with their importances ? Please see picture below - the features are quite smushed together and I would like to present them in a different manner or not present them at all.

The text was updated successfully, but these errors were encountered: