-
Notifications
You must be signed in to change notification settings - Fork 845
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Error breaking StereoBonds in reactions #3147
Comments
|
Interesting, thanks. So the main difference seems to be the version? |
You could try to run the code in the different version. |
|
This does look like a bug, but it isn't with the components, it appears to be with chirality: This fails: This passes: |
|
@greglandrum Chirality issues should get auto assigned to you :) |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Version '2019.09.3'
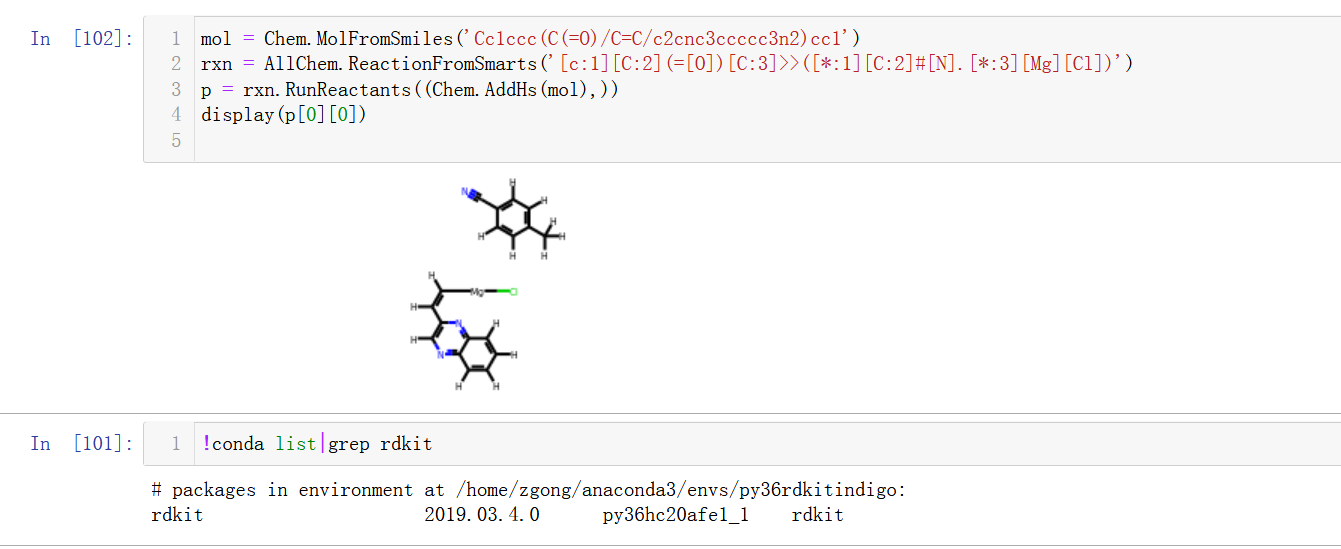
I am just noting this strange behavior where the intramolecular reaction does not seem to be allowed when disconnecting a bond to an atom that is part of a separate double bond.
Example to reproduce
gives
Though removing the outer parentheses to give
rxn = AllChem.ReactionFromSmarts('[c:1][C:2](=[O])[C:3]>>[*:1][C:2]#[N].[*:3][Mg][Cl]')makes the reaction work perfectly fine. I am somewhat confused by this and appreciate any help with the issue. Thanks!The text was updated successfully, but these errors were encountered: