This repository contains matlab code to:
- Project 3D volumetric nifti images in 2mm isotropic MNI space (see example file in folder 'volumetric') to the HCP MMP surface
- Extract values of each parcel in the Glasser atlas
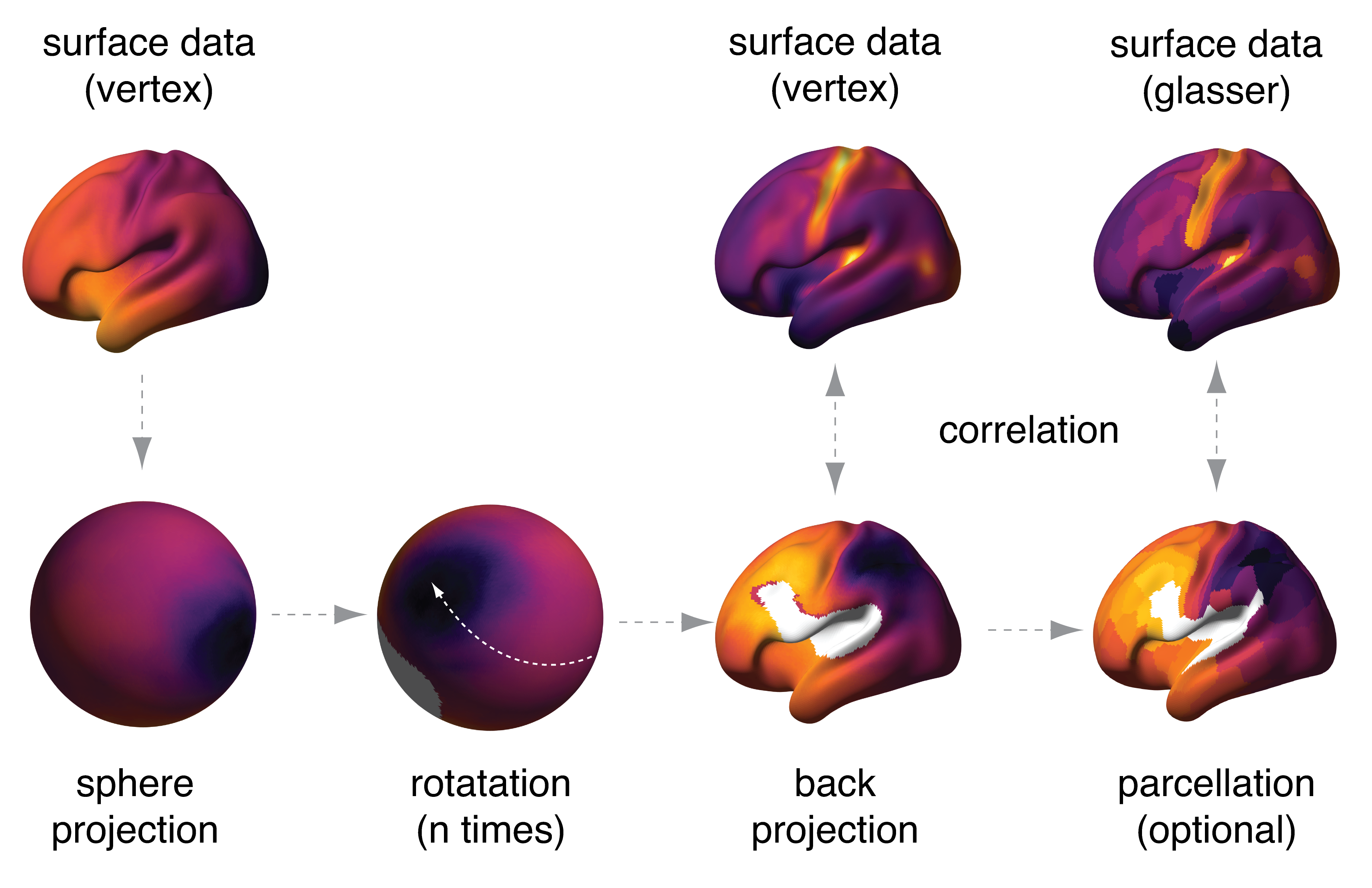
- Create null maps by spherical rotation
- Test spatial correlation between maps and compute p-values that take into account spatial autocorrelation in the maps via spherical rotation ("spin testing").*
See example.m for syntax.
This code will not run out of the box. You'll need:
- An installed version of the human connectome workbench software (https://www.humanconnectome.org/software/get-connectome-workbench)
- Fieldtrip (http://www.fieldtriptoolbox.org/)
- To edit the relevant lines in the "functions/pathfindr();" so that these packages are found by matlab (lines 21, 25, and possibly line 23 depending on your OS)
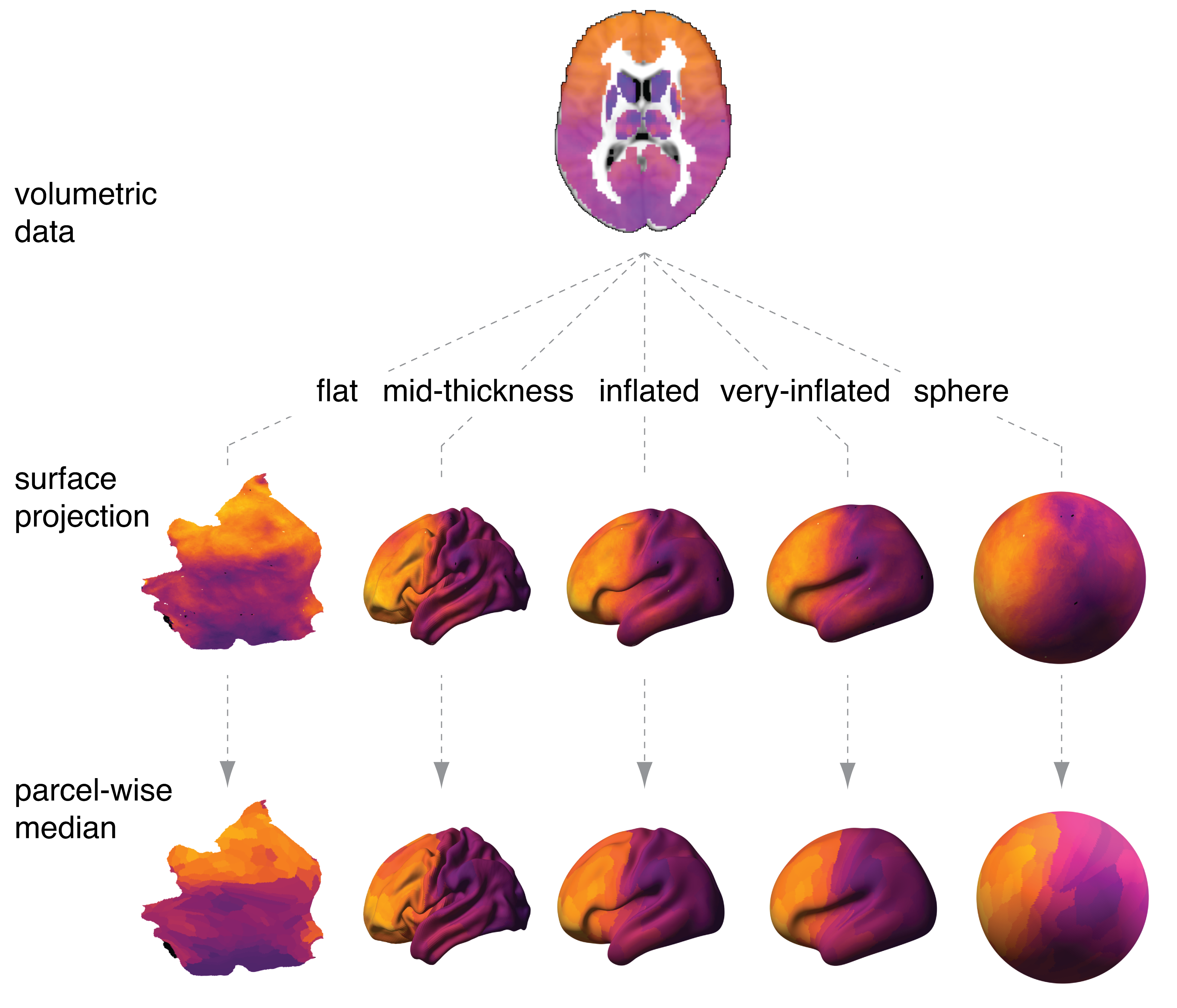
Surface projection will allow you to do the following:
Spin testing works as follows:
Correlate_surface(); does the spin testing. It requires two input arguments, of which at least one is vertex-level data. In case one of the input surfaces is pre-parcellated, use the 'glasser' option. In this case, use the parcellated map as the first input, and the vertex-level map as the second input. The medial wall of the original surface as well as the rotated surrogate surfaces (white parts in the figure above) are blanked out and excluded from correlation, so each correlation in the null distribution will have a slightly different number of parcels / vertices that are included.
Code to compute and plot parcel-wise median values of a T1 / T2 contrast (myelin map) is also provided. The myelin map as well as the surface files are from the HCP S1200 release (https://www.humanconnectome.org/study/hcp-young-adult/document/1200-subjects-data-release). The file 'mvals.csv' also contains the parcel-wise median values of the myelin map. The file 'tvals.csv' contains parcel-wise median values of a cortical thickness map.
Labels to atlas parcels can be found in Glasser_labels.csv.
List of functions:
- glasserize_nifti: produce parcel-wise median values for a volumetric nifti (.nii) file
- glasserize_cifti: produce parcel-wise median values for a surface file
- glasserize_surface: produce parcel-wise median values for a surface vector
- surface_project: project a volumetric file to the cortical surface, and return spatially z-scored vertex-wise values
- surface_project_raw: project a volumetric file to the cortical surface, and return the raw vertex-wise values
- correlate_surface: correlate two surface vectors while controlling for spatial autocorrelation
- sphere_rotate: produce spherically rotated surrogate maps
- cortsurfl: plot data onto the cortical surface of the left hemisphere
- cortsurfr: plot data onto the cortical surface of the right hemisphere
- pathfindr: function to set paths
- inferno: the inferno color map
References:
- Alexander-Bloch, AF, Shou, H, Liu, S, Satterthwaite, TD, Glahn, DC, Shinohara, RT, Vandekar, SN, Raznahan, A, (2018) On testing for spatialcorrespondence between maps of human brain structure and function. Neuroimage 178, 540-551. doi:10.1016/j.neuroimage.2018.05.070