New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Difficulty in Setting Initial Conditions #7
Comments
|
Hi - thanks for your notes. It is certainly not what I expect. Is it possible to share your code? You can email me privately and I will check that out. |
|
Sure, here is the file. Thank you for your reply. I tried modifying a few of the conditions - I am just surprised that the loss stays completely constant over the epochs and there is no evidence it picked up the initial condition at all (just flat everywhere). Especially when the linear one with the discontinuous boundary does work. |
|
I spot an issue in the C1:
This relation does not impose any condition on the network (rho). Therefore it does not change at all as there is no parameter gets affected by this condition. Is it possible to have a typo in C1, i.e. |
Hello,
I have been trying to write a code to solve a reaction-diffusion type problem. I wanted to compare deepXDE and SciANN but was having difficulty getting my initial conditions set up in SciANN.
In deepXDE, I can use
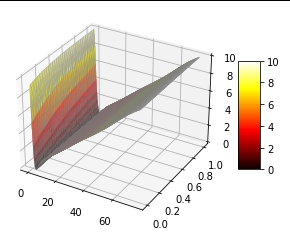
rho0*tf.cast(tf.math.greater(((x-x_center_scaled)**2)/(x_axis_scaled**2),1),tf.float64)To set a circle (line segment in 1D for now) in the center of the domain to zero while keeping everything else at a constant value. The result is shown below (sorry for the unlabeled axes - that is x from 0 to 75 and t from 0 to 10).
In SciANN, this doesn't seem to work since the Functionals cannot be acted on by TF operations. In accordance with the Burger eq example, I tried using
0.25*(1 - sign(tt - TOL)) * ((1 - sign(x - (left_boundary+TOL))) + (1 + sign(x - (right_boundary-TOL)))) * ( rho0)as well as
0.5*(1 - sign(t - TOL)) * rho0 * sign(((x-x_center)**2)/(x_axis**2))In numpy, both of these give the correct shape
I also tried using
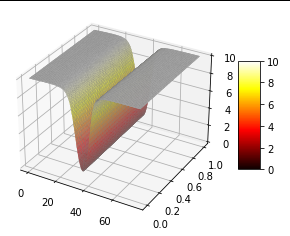
0.5*(1 - sign(t - TOL)) * rho0 * tanh(((x-x_center)**2)/(x_axis**2))Which should give a smoother boundary. In all cases the whole domain appears as solid rho0, with no area of 0 in the center, as shown below.
I also note that the initial condition loss seems to immediately get stuck at a particular value.
So I am wondering: is there a problem with the way I have set up my boundary conditions, a way to use TF operations or a "greater_than" function in SciANN, or is there another way to set up my boundary conditions?
Thanks,
David
The text was updated successfully, but these errors were encountered: