New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
More flexible fitting function, allow likelihood, remove uncertainties dependency #149
Conversation
|
Can you verify my changes are valid? Could you add a test with pytest-mpl, perhaps? |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Notes on my changes.
| @@ -45,7 +46,7 @@ def _filter_dict( | |||
| } | |||
|
|
|||
|
|
|||
| def _expr_to_lambda(expr: str) -> Callable: | |||
| def _expr_to_lambda(expr: str) -> Callable[..., Any]: | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is the default for Callable - best to not leave empty Generics.
| ydata: ArrayLike, | ||
| yerr: ArrayLike, | ||
| func: Callable[..., Any], | ||
| xdata: np.ndarray, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ArrayLike can be a simple number, so xdata[stuff] was not valid. Either use np.asarray, or just require arrays.
| likelihood: bool = False, | ||
| ) -> Tuple[ArrayLike, ArrayLike]: | ||
| ) -> Tuple[Tuple[float, ...], ArrayLike]: |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Makes the typing easier, because NumPy's typing is a bit spotty. It doesn't know that *array is valid, etc. This is small so was a simple fix.
src/hist/plot.py
Outdated
| p0 = None | ||
| if func.__defaults__ and len(func.__defaults__) + 1 == func.__code__.co_argcount: | ||
| p0 = func.__defaults__ | ||
| params = list(inspect.signature(func).parameters.values()) | ||
| p0 = [ | ||
| 1 if arg.default is inspect.Parameter.empty else arg.default | ||
| for arg in params[1:] | ||
| ] | ||
|
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm using inspect.signature, as it's a more public interface; __defaults__ (and maybe __code__) are untyped, AFAICT (or maybe just not always present on Callables). This also supports partial defaults, while the other was all or nothing.
| from iminuit import Minuit | ||
| from scipy.optimize import curve_fit | ||
| from iminuit import Minuit # noqa: F401 | ||
| from scipy.optimize import curve_fit # noqa: F401 | ||
| except ImportError: |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This could be a ModuleNotFoundError.
src/hist/plot.py
Outdated
| if type(func) in [str]: | ||
| if isinstance(func, str): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Never check the type exactly, always use isinstance. It supports subclassing and MyPy type narrowing.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also never make a list for containership testing, use a set, it's faster. x in {a, b}.
src/hist/plot.py
Outdated
| constant = ydata.max() | ||
| constant = float(ydata.max()) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Not always fond of NumPy's choices here. Not sure what a number[Any] is. So just forcing it to a Python float.
| parnames = func.__code__.co_varnames[1:] | ||
| assert not isinstance(func, str) | ||
|
|
||
| parnames = list(inspect.signature(func).parameters)[1:] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Same higher level usage of inspect.signature over .__code__.co_varname.
|
@all-contributors please add @aminnj for code |
|
I've put up a pull request to add @aminnj! 🎉 |
|
Also, would you like to fill in the changelog so we can clear that needs-changelog badge? :) (I can do it if you prefer) |
|
I appreciate the comments. I haven't yet grasped all the subtleties of the type system... Is the suggestion to use |
|
This also need docs, but don't worry about that - I'll be reworking the docs in the near future to make them easier to edit. When I do, I'll add to this part. There are two ways to test. The easier way is to use The better way to test is to mock mpl and then verify the sequence of commands to the mpl functions do not change. That also gives much better error messages if something gets changed, rather than just an image diff. But that's much harder to set up. You can see what I did for mplhep here: https://github.com/scikit-hep/mplhep/blob/5effa0b857d7eea93da0299cbbcdc777bd3abaaf/tests/test_mock.py - but as you see, I didn't end up adding it for everything because it's a bit of work for each one. |
for more information, see https://pre-commit.ci
for more information, see https://pre-commit.ci
|
Looks good to me, can you add a changelog entry, then we can merge? If not, let me know, and I'll add one; I usually lightly push contributors to add changelog entries but add them if not; original contributors are usually the best at advertising what they have done. :) |
|
I added in some test cases that I had forgotten earlier (the |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks great, thanks!
|
Hi @henryiii, @aminnj, I'm following your nice developments. I just wanted to make a comment now that I see |
|
I personally have no strong attachment to "gaus", so I can offer a +1 to "gauss" |
|
I was taking a look at https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.rv_continuous.html today and noticed it has a |
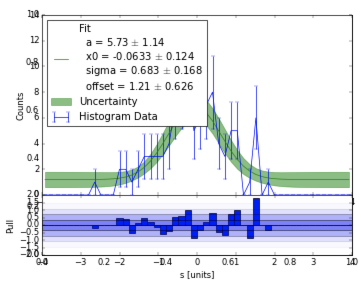
Based on the discussion in #146, I added some features to
plot_pull. The theme is making fitting more streamlined for exploration.curve_fitinitial guess (p0) from default arguments, if they existplot_pull("a+b*x"))plot_pull(..., likelihood=True)) (chi2 by default, as before)uncertainties.numpydependency and construct band by resampling covariance matriximinuit(gets the covariance matrix right, unlikescipy.optimizemost of the time), but the initial guesses foriminuitare seeded fromscipySetup
Before (including a bug-fix for the
variancesfrom the above issue):After: