New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
how to extract the most important feature names ? #632
Comments
|
The array returned by |
|
I understand that clarifyingMy issue, is about getting the names and their shap values of features, instead of visualizing them. exampleTake the demo in shap.summary_plot(shap_values, X, plot_type="bar")However, I am wondering, is there any way to get the name of features ordered by importance: # is there any function working like get_feature_importance? Or how to implement it?
shap.get_feature_importance(shap_values, X) == np.array(['LSTAT', 'RM', 'CRIM', ... 'CHAS']) Or even better, output the numeric values of each feature: # is there any function working like get_feature_importance_2? Or how to implement it?
shap.get_feature_importance_2(shap_values, X) == {
'LSTAT': 2.6,
'RM': 1.7,
...,
'CHAS': 0.0
}Thank you so much. |
|
Ah. Well the numbers for the bar chart are just |
|
I usually do this to get feature importance. vals= np.abs(shap_values).mean(0)
feature_importance = pd.DataFrame(list(zip(X_train.columns,vals)),columns=['col_name','feature_importance_vals'])
feature_importance.sort_values(by=['feature_importance_vals'],ascending=False,inplace=True)
feature_importance.head() |
I think the shap_values format has changed. import numpy as np
vals= np.abs(shap_values).mean(0)
feature_importance = pd.DataFrame(list(zip(features.columns, sum(vals))), columns=['col_name','feature_importance_vals'])
feature_importance.sort_values(by=['feature_importance_vals'], ascending=False,inplace=True)
feature_importance.head() |
|
I think it would be useful to have this as a "feature_importance()" method, which I will look at adding as a pull request if anyone agrees? On a different note, but related to this topic... I have only read summary information on "shap" and from my understanding, the shap_values are related to the individual observations (not at the global level) as such, I wondered how to use for an obscure cross validation situation I have... I would like to find feature importances based on the training data of all the folds during CV (fully stratified folds, so no danger of missing variables/values in a particular fold etc). I wondered if I should:
|
|
if the feature importance does not adds upto one ? is it incorrect? |
For anyone seeing this more recently, even if you are doing a binary classification problem, the returned To reflect that in the code snippet above, I had to specifically select the positive class in my case in order to land on correct shap values: import numpy as np
vals= np.abs(shap_values[1]).mean(0) |
|
For anyone who are looking for a solution in Shap version 0.38.1, I can reproduce the shap importance plot order by taking the absolute value of the proposed solution of @clappis. "features" is the list of features, you can use validation.columns instead.
|
|
For Shap version 0.39 : def global_shap_importance(model, X):
""" Return a dataframe containing the features sorted by Shap importance
Parameters
----------
model : The tree-based model
X : pd.Dataframe
training set/test set/the whole dataset ... (without the label)
Returns
-------
pd.Dataframe
A dataframe containing the features sorted by Shap importance
"""
explainer = shap.Explainer(model)
shap_values = explainer(X)
cohorts = {"": shap_values}
cohort_labels = list(cohorts.keys())
cohort_exps = list(cohorts.values())
for i in range(len(cohort_exps)):
if len(cohort_exps[i].shape) == 2:
cohort_exps[i] = cohort_exps[i].abs.mean(0)
features = cohort_exps[0].data
feature_names = cohort_exps[0].feature_names
values = np.array([cohort_exps[i].values for i in range(len(cohort_exps))])
feature_importance = pd.DataFrame(
list(zip(feature_names, sum(values))), columns=['features', 'importance'])
feature_importance.sort_values(
by=['importance'], ascending=False, inplace=True)
return feature_importance |
|
shap==0.39.0 In my case the above code didn't work, but gave and idea what to do. I had an xgboost model and did: explainer = shap.Explainer(model) shap_values.shape => (5, 4000, 30) (5 instances, 4000 features, 30 classes) vals = shap_values.values And finally some numbers looking like feature importances popped out. |
|
Hey, I have a very similar question. I am interested in exporting the data used to create the shap.dependence_plot("rank(0)", shap_values, X_train) diagrams. Anyone could help me with that? I basically wanne save them in an excel sheet, to plot them bymyself. thx |
|
How do we get a list of the most important features for a particular class in a multiclassification problem? My question is: Using the array corresponding to each class (eg: For class 0, using array shap_values[0] ), how can I get a list of most important features for that particular class? TLDR: Basically, I want to save the features shown on the y-axis (AT1G09740.1, etc) from the summary plot in a txt file. How do I do that? |
I think I figured out the answer. This is what I did for Class 0: |
Now it wants vals = np.abs(shap_values.values).mean(0)
feature_importance = pd.DataFrame(list(zip(feature_names, vals)), columns=['col_name','feature_importance_vals'])
feature_importance.sort_values(by=['feature_importance_vals'], ascending=False, inplace=True)
feature_importance.head() |
|
` X, y = shap.datasets.boston() explainer = shap.Explainer(model) feature_names = list(X.columns.values) ` |
This function works like a charm! Thanks! |
|
feature_importance = pd.DataFrame(list(zip(feature_names, np.abs(shap_values)[0].mean(0))), columns=['feature_name', 'feature_importance_vals']) Before, you are putting np.abs(shap_values[0]).mean(0)), and this is no correct because you would be selecting the first row and doing the mean. The correct way is: np.abs(shap_values)[0]. |
|
Version 0.40.0: Verified the same as summary bar plot |
|
If anyone is still wondering, after sorting through the code for a bit, I realized the solution is incredibly simple (This is with XGBoost. For LightGBM, Confirmed the order is the same as the summary_plot. |
|
I am very curious why there is no example as to how to extract the feature importance of lagged shap values (shape (22, 2, 9)) returned by a DeepExplainer. Any method i tried did not return the same order and impact as for example summary plot did. I would appreciate a solution for my issue as i am stuck to summarize feature importance for my thesis |
|
I had to use |
|
How can i see the detailed description of features? Which function to use? school object |
|
@slundberg I'm curious as to why this is not already implemented as a feature. Any particular reason, or difficulty you foresee? Or there just hasn't been anyone who creates the PR? Thank you |
|
I had exactly the same issue today and found a fairly easy way to get the top n features by shap value: then just pick off your top n features from the head of the df_feature_importance dataframe. I'm not a python expert, so I have no doubt there's a more elegant way of doing this, but it solves the issue. |
With this code we have importance in Series |
Did you find any solution? Please tell me |
|
@slundberg, have something like UPD: |

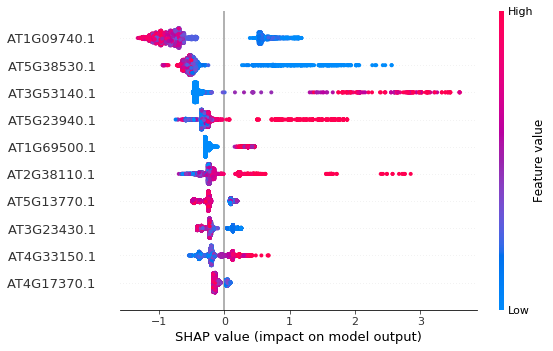

We can visualize the feature importance by calling
summary_plot, however it only outputs the plot, not any text.Is there any way to output the feature names sorted by their importance defined by
shape_values?Something like:
Thanks.
The text was updated successfully, but these errors were encountered: