OOP Perl module to convert Sanger trace files (.ab1) to FASTQ sequences. Ambiguities (hetero bases) are managed.
Homepage: FASTX::Abi (MetaCPAN)
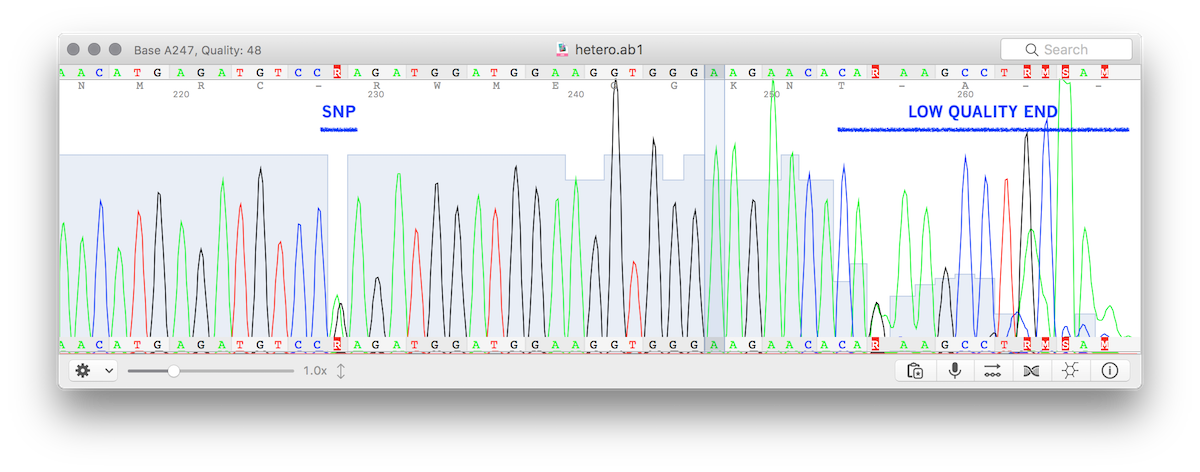
The picture shows a trace file (chromatogram, in abi/ab1 format) with a SNP detected and the typical quality degradation at the end.
Via cpanminus:
# Install cpanminus if you don't have it:
curl -L https://cpanmin.us | perl - --sudo App::cpanminus
# Install FASTX::Abi
cpanm FASTX::Abi
Via Miniconda:
# Install FASTX::Abi
conda install -y -c bioconda perl-fastx-abi