Using a convolutional neural network, transfer learingn and deep neural network attribution methods to localize abnormalities on x-ray chest images.
The project is right now in its initial stage. It still requires some fine tuning. I've managed just to put all the pieces together for it to work.
Special thanks are due to:
- Ayush Singh (@ayush1997). After making some small changes, I've reused his scrapper to download x-ray images from the openi.nlm.nih.gov website (https://github.com/ayush1997/Xvision). My project is also inspired by the way Ayush uses the VGG16 CNN to classify x-ray images.
- Marco Ancona (@marcoancona) for his amazing library called DeepExplain (https://github.com/marcoancona/DeepExplain). It provides (used in this project) attribution methods for Deep Learning that are compatible with tensorflow.
The VGG16 is a base of the classification model used to distinguish between normal and abnormal x-ray images. It has been stripped of all fully connected layers. These layers have been replaced with new ones, trained with a transfer learning techniques.
| Precision | Recall | F-Score | Support | |
|---|---|---|---|---|
| Normal | 0.58 | 0.56 | 0.57 | 259 |
| Abnormal | 0.77 | 0.79 | 0.78 | 488 |
| avg/total | 0.71 | 0.71 | 0.71 | 747 |
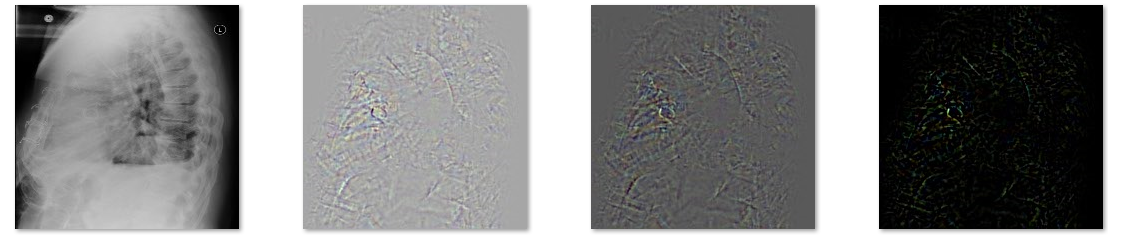
Abnormalities are located using DeepLIFT attribution method.
To run the whole process you have to follow these steps. (You might have to create data and data/model paths manually)
python scraper/download_model.py <vgg_path>
e.g.
python scraper/download_model.py data/vgg16.tfmodel
python scraper/scraper.py <path>
e.g.
python scraper/scraper.py data
python learning/transfer_feature_extraction.py <images> <features> <vgg_path>
e.g.
python learning/transfer_feature_extraction.py data/images data/transfer_features.pickle data/vgg16.tfmodel
In this step vgg16 network is split and only convolution layers are used.
python learning/learning_examples_preparing.py <descriptions> <features> <training_set> <testing_set> <examples_list>
e.g.
python learning/learning_examples_preparing.py data/images-description.json data/transfer_features.pickle data/training.pickle data/testing.pickle data/examples.json
It produces training and testing examples used in the training process.
python learning/fully_connected_layers_training.py <training_set> <testing_set> <model>
e.g.
python learning/fully_connected_layers_training.py data/training.pickle data/testing.pickle data/model/model.ckpt
Features that have been previously extracted from convolutional layers are now used to train fully connected layers.
python learning/visualization.py <vgg_path> <model> <examples_list> <images> <results> <attribution_method>
e.g.
python learning/visualization.py data/vgg16.tfmodel data/model/model.ckpt data/examples.json data/images data/results deeplift
Examples on <examples_list> are now passed through VGG16 CNN connected with our fully connected layers. DeepLIFT attribution method is then used to localize abnormalities. Results are sroted in <results> directory. Names of output files contain information about prediction [normal probability, abnormal probability].