New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ValueError when writing 2D seismic data to file after reprojecting coordinates #109
Comments
|
So I've downloaded your data and tried to run an import export workflow. The following appears to work for me Can you try removing relative paths, ensure the call to This is what the loaded data looks like for me: |
|
Ok, Sorry, I didn't see the notebook. This cell is your issue When you assign numpy arrays back to and xarray Dataset it doesn't know how to interpret the dimension of what you are assigning. The assign operation overwrites any variable that was there before, so previous variables are ignored. What xarray does when it doesn't have the dimension specified is it creates a new dimension with the variable name. So after this operation You can fix this two ways, specify the dimensions in the assignment Or access the data array directly by indexing |
|
@trhallam I finally managed to convert the coordinates :) I will implement the functionality in GemGIS as it is quite useful for us working with cross-country seismic. I will also reference a notebook here later. |
|
@trhallam I noticed something weird: |
|
Possibly, you might have to set a coordinate scaler in the attributes of the Dataset before you write it out. If you load a 2d line that someone else has created in the same area, you'll probably see a scalar like -100. |
|
The coordinates actually fit when reading the data. How would you set the coordinate scalar? |
|
The problem is not so much the Python side but the SEGY side, there will never be an issue reading in the data, and the coordinates are transformed using the scalar from the trace headers. The trace header standard in SEGY limits the size of a number to 4 bytes. So the biggest number you can store is a 16 bit float I think, while floats in Python are typically 32 or 64 bit. This limits the maximum number of exponent bits you can store. Thus moving them into the fraction component reduces accuracy but typically we don't need more than 2 or 3 decimals for coordinates. Sub-mm accuracy isn't practical for most seismic. To write the data out you should set the before you write out the data. The scalar follows the SEGY conventions where -100 would mean You can see if this attribute is set to something on the data which you read in previously. |
|
Yes, that worked! It was not
but
that did the trick. Thanks for the prompt help! I hope everything works now :D |
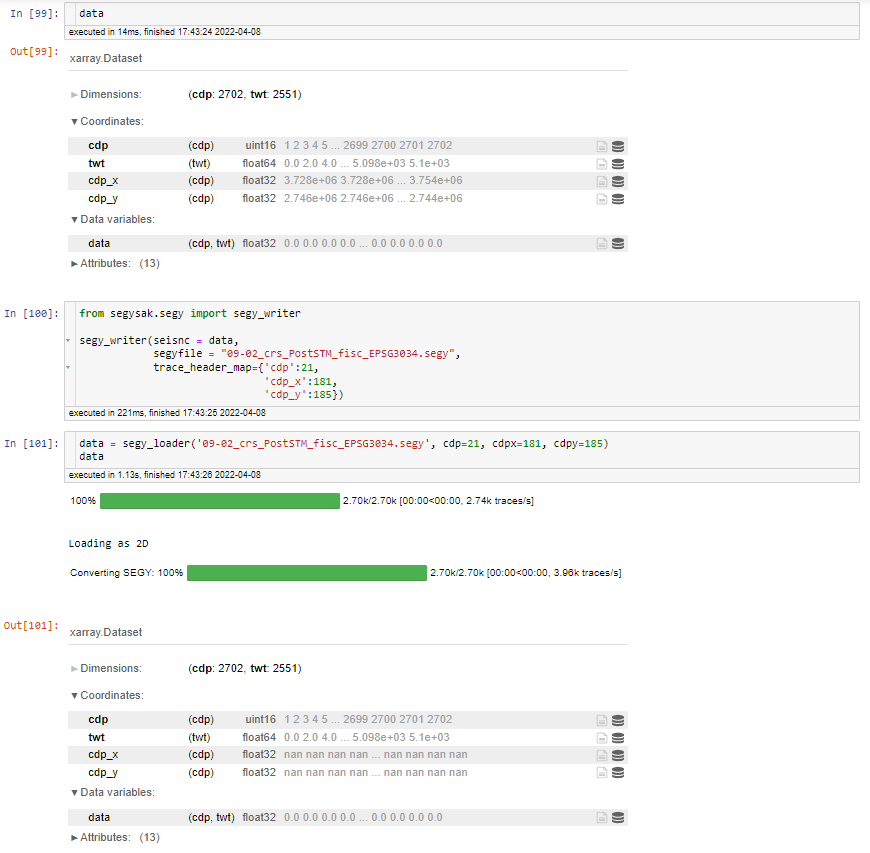


Hello,
I am receiving a
ValueErrorwhen replacing the original CDP coordinates with reprojected coordinates and then trying to save the data set again.I have attached the notebook and the data, the data can be freely downloaded from https://www.nlog.nl/index.php/en/node/588
The error occurs when trying to save the data:
Here is the full error message:
09-02_crs_PostSTM_fisc.sgy.zip
[09-02_crs_PostSTM_fisc.sgy.zip]
The text was updated successfully, but these errors were encountered: