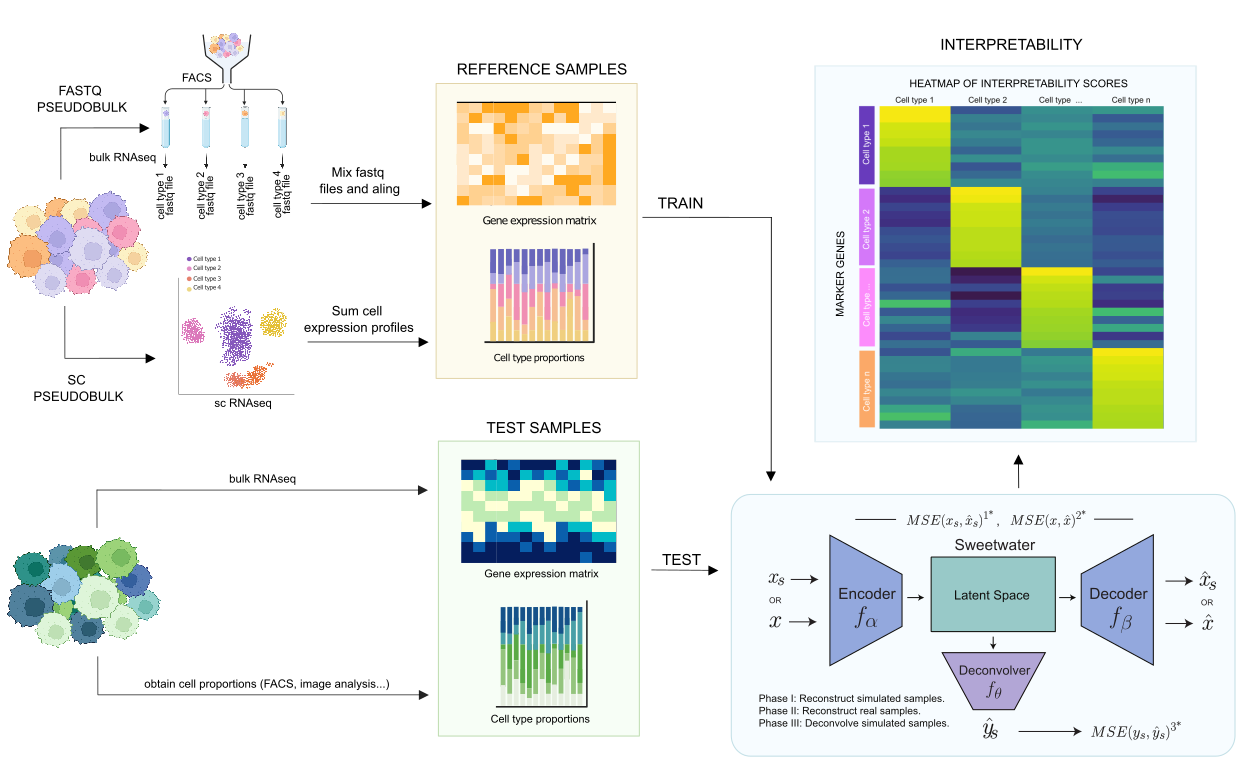
Single-cell RNA-sequencing (scRNA-seq) stands as a powerful tool for deciphering cellular heterogeneity and exploring gene expression profiles at high resolution. However, its high cost renders it impractical for extensive sample cohorts within routine clinical care, hindering its broader applicability. Hence, many methodologies have recently arised to estimate cell type proportions from bulk RNA-seq samples (known as deconvolution methods). However, they have several limitations:
- Many depend on selecting a robust scRNA-seq reference dataset, which is often challenging.
- Secondly, building reliable pseudobulk samples requires determining the optimal number of genes or cells involved in the simulated data generation process, which has not been studied in depth. Moreover, pseudobulk and bulk RNA-seq samples often exhibit distribution shifts.
- Finally, most modern deconvolution approaches behave as a black box, and the underlying mechanisms of the deconvolution task are still unknown, which can compromise the reliability of the results.
In this work, we present Sweetwater, an adaptive and interpretable autoencoder able to efficiently deconvolve bulk RNA-seq and microarray samples leveraging multiple classes of reference data, such as scRNA-seq and single-nuclei RNA-seq. Moreover, it can be trained on a mixture of FACS-sorted FASTQ files, which we newly propose to use as this reduces platform-specific biases and may potentially outperform single-cell-based references. Also, we demonstrate that Sweetwater effectively uncovers biologically meaningful patterns during the training process, increasing the reliability of the results.
In order to use Sweetwater, we have included an example.ipynb file where it is showed how to deconvolve an expression matrix using a scRNA-seq reference, perform the interpretability analysis showed in the manuscript, and generate pseudobulk samples using FASTQ references.