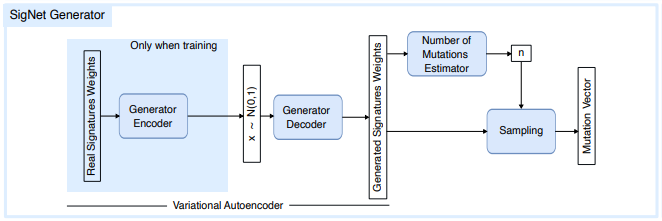
SigNet Generator is composed of three modules sequencially called:
-
Variational auto-encoder (VAE): This network is capable of generating realistic-looking signature compositions. Variational auto-encoders are deep generative models composed of two blocks: an encoder and a decoder. The encoder non-linearly projects the inputs into a latent space of a pre-fixed dimension, while the decoder recovers the original input from this projection.
When sequentially applying encoder and decoder, one obtains an approximation of the identity function.
What makes them particularly useful in our case is the application of some conditions in the latent space.
For instance, we force the projected data to follow a p-dimensional Gaussian
$\mathcal{N}(0, 1)$ distribution. Therefore, we obtain a mapping between the original data distribution and a standard multidimensional Gaussian. This allows us to generate realistic-looking mutational vectors, simply by sampling from a Gaussian and serving those samples as inputs to the decoder. Effectively, this network is learning the underlying correlations between signatures, being capable of generating previously unseen but plausible examples. In our case, both the encoder and the decoder are modeled by feed-forward ANNs, composed of standard linear layers with leaky ReLU activation. It was trained using PCAWG dataset as input/labels. Furthermore, we use the re-parametrization trick for variance reduction: while training, given an input$x_i$ from the dataset$\mathcal{D} = { x_i }_{i=1:N}$ , where$N$ denotes the training dataset size, the encoder returns two values:$\mu_i$ and$\sigma_i$ . These values are then used to sample the input from the decoder$z_i \sim \mathcal{N}(\mu_i, \sigma_i)$ , where$\mathcal{N}(\mu,\sigma)$ denotes the normal distribution with mean$\mu$ and standard deviation$\sigma$ . Finally, the decoder takes this sample$z_i$ and returns a reconstruction$\hat x_i$ .
Feeding values from a standardized normal, i.e.
-
Number of Mutations Estimator Network: This module is an ANN trained to predict the number of mutations range of a given signature composition vector. Different cancer types present different mutational counts. It is important to correctly pair the generated weights and its associated number of mutations.
-
Sampling algorithm: Given a signature catalog, a signature composition vector and a certain number of mutations, this block is a simple sampling algorithm that generates a mutational vector.
This algorithm presents some trade-offs when compared to the current state-of-the art SynSigGen. Its main advantage being that it encodes the main mutation correlations existent in the training data. SynSigGen instead, independently attributes weight to signatures depending on their frequencies, thus ignoring those correlations. It is important to notice, however, that spurious correlations are created by our method. Further work should be done on a model level to address them. Check out the manuscript for further analysis.