Table of Content
The small demo with 359k spectra is using default port 80 now.
Demo 1: a demo archive with 25 data files, ~359,022 spectra http://spectroscape.cc
Demo 2: an archvie with 25 million spectra. http://spectroscape.cc:8709
Demo 3: an archive with > 100 million spectra. http://omics.ust.hk:8709
Feb 19. 2024We created our first DOI badge from Zenodo. Future releases may also come with DOI links.
Currently, Zenodo release (as well as GitHub source code package) does not contain the web UI submodule. Therefore, please get our newest source code from GitHub release page to test. Please refer to the Source code installation part for detailed steps.
Sep. 13, 2023
Now there are three demos available.
Demo 1: a demo archive with 25 data files, ~359,022 spectra http://spectroscape.cc
Demo 2: an archvie with 25 million spectra. http://spectroscape.cc:8709
Demo 3: an archive with > 100 million spectra. http://omics.ust.hk:8709
Aug 23. 2023 in HKUSTOur server now is switched to use default port 80. Please visit: http://spectroscape.cc for an demo spectral archive. We temporarily turn off the demo on port 8709.
July 31, 2023Our server in HKUST was shut down during the weekend due to a electricity suspension. Now the spectroscape demo is back online.
Please visit http://spectroscape.cc:8709 for an demo spectral archive with over 25 million spectra.
June 26, 2023 in HKUSTWe made our spectral archive search demo available on the following website.
http://spectroscape.cc:8709
We will keep this domain in the near future and make our Spectroscape available to everyone.
June 23, 2023
Spetroscape is a software tool to search for similar PSMs in spectral archives. It can create a spectral archive, and incrementally add new data (mzML/mzXML) and annotations (pep.xml) to it.
Spectroscape has a web user interface, which enables real time searching for approximate nearest neighbors (ANNs) against an archive with hundreds of millions of spectra.
For installation, users may either follow the YouTube video tutorial below or read through the next section of the ReadMe.md file.
If you would like to use Spectroscape directly via brower, click here.
This following command has been tested on Ubuntu 22.04 and 20.04. The .deb file of latest version of spectroscape can be found in the following link.
Spectroscape comes with both CPU and GPU versions. If CUDA environment is not available, please use CPU version.
Spectroscape (CPU version) can be installed using following command lines.
wget https://github.com/wulongict/SpectralArchive/releases/download/v1.1.9/Spectroscape_CPU-1.1.9.deb
sudo apt update
sudo apt install ./Spectroscape_CPU-1.1.9.deb
In case that the user do not have root privilege, the following command could be used.
wget https://github.com/wulongict/SpectralArchive/releases/download/v1.1.9/Spectroscape_CPU-1.1.9.deb
dpkg -x ./Spectroscape_CPU-1.1.9.deb ./The GPU version can be installed similarly.
wget https://github.com/wulongict/SpectralArchive/releases/download/v1.1.9/Spectroscape_GPU-1.1.9.deb
sudo apt update
sudo apt install ./Spectroscape_GPU-1.1.9.debHowever, users should first make sure CUDA environment available. Otherwise, the following error occurs when running spectroscape.
spectroscape
spectroscape: error while loading shared libraries: libcudart.so.11.0: cannot open shared object file: No such file or directoryuse the following command line to remove spectroscape (both GPU and CPU versions) from Ubuntu system.
sudo apt remove spectroscape_cpu spectroscape_gpuThis following command has been tested on Ubuntu 22.04. It does not compile on older versions because of
the cmake_minimum_required parameter.
cmake and gcc are required to compile of C++ code.
sudo apt update
sudo apt install cmake build-essential The source code requires libopenblas.
sudo apt install libopenblas-dev To make the web interface work, two more tools should be installed, spawn-fcgi and nginx.
sudo apt install spawn-fcgi nginxFinally, to compile GPU version, CUDA environment is required.
First, get the latest source code of spectroscape from GitHub.
git clone --recurse-submodules https://github.com/wulongict/SpectralArchive.git --branch releaseStart from here, all the command should be executed under the source code folder, namely, SpectralArchive.
Run the following scripts to remove any intermediate files and have a clean start.
./cleanMake.bashUsers can compile a CPU or GPU version using option FALSE or TRUE.
# CPU version
./compile.bash FALSE# GPU version
./compile.bash TRUEAfter the compilation, the executable files are under the build/bin folder inside the source code directory.
build/
├── bin
├── include
├── lib
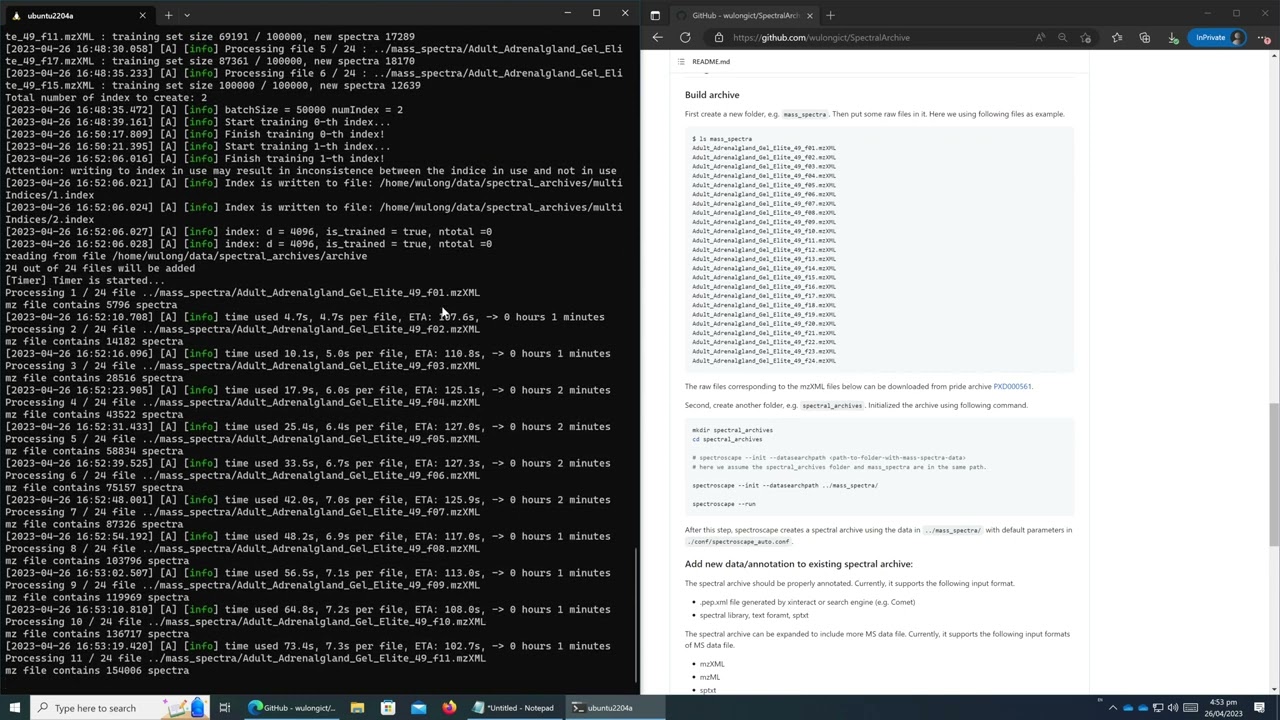
└── shareFirst create a new folder, e.g. mass_spectra. Then put some raw files in it. Here we using following files as example.
Note that the minimum number of spectra required to initialize a spectral archive by Spectroscape is 100,000. Using only
one mzXML file is not adquate to build an archive. Please try download the following files
from this link to Google Drive.
Users could also
download the compressed version
from Google Drive.
$ ls mass_spectra
Adult_Adrenalgland_Gel_Elite_49_f01.mzXML
Adult_Adrenalgland_Gel_Elite_49_f02.mzXML
Adult_Adrenalgland_Gel_Elite_49_f03.mzXML
Adult_Adrenalgland_Gel_Elite_49_f04.mzXML
Adult_Adrenalgland_Gel_Elite_49_f05.mzXML
Adult_Adrenalgland_Gel_Elite_49_f06.mzXML
Adult_Adrenalgland_Gel_Elite_49_f07.mzXML
Adult_Adrenalgland_Gel_Elite_49_f08.mzXML
Adult_Adrenalgland_Gel_Elite_49_f09.mzXML
Adult_Adrenalgland_Gel_Elite_49_f10.mzXML
Adult_Adrenalgland_Gel_Elite_49_f11.mzXML
Adult_Adrenalgland_Gel_Elite_49_f12.mzXML
Adult_Adrenalgland_Gel_Elite_49_f13.mzXML
Adult_Adrenalgland_Gel_Elite_49_f14.mzXML
Adult_Adrenalgland_Gel_Elite_49_f15.mzXML
Adult_Adrenalgland_Gel_Elite_49_f16.mzXML
Adult_Adrenalgland_Gel_Elite_49_f17.mzXML
Adult_Adrenalgland_Gel_Elite_49_f18.mzXML
Adult_Adrenalgland_Gel_Elite_49_f19.mzXML
Adult_Adrenalgland_Gel_Elite_49_f20.mzXML
Adult_Adrenalgland_Gel_Elite_49_f21.mzXML
Adult_Adrenalgland_Gel_Elite_49_f22.mzXML
Adult_Adrenalgland_Gel_Elite_49_f23.mzXML
Adult_Adrenalgland_Gel_Elite_49_f24.mzXML
The raw files corresponding to the mzXML files below can be downloaded from pride archive PXD000561.
Second, create another folder, e.g. spectral_archives. Initialized the archive using following command.
mkdir spectral_archives
cd spectral_archives
# spectroscape --init --datasearchpath <path-to-folder-with-mass-spectra-data>
# here we assume the spectral_archives folder and mass_spectra are in the same path.
spectroscape --init --datasearchpath ../mass_spectra/
spectroscape --run
After this step, spectroscape creates a spectral archive using the data in ../mass_spectra/ with default parameters
in ./conf/spectroscape_auto.conf.
The spectral archive can be expanded to include more MS data file. Currently, it supports the following input formats of MS data file.
- mzXML
- mzML
- sptxt
The spectral archive should be properly annotated. Currently, it supports the following input format.
- .pep.xml file generated by xinteract or search engine (e.g. Comet)
- spectral library .sptxt
- text format .spectroscape.tsv
For the last tsv format, please follow the example file in tests/data/example.spectroscape.tsv. Here is how the format
looks like. The first line is header and should not be changed.
filename scan modpep charge protein ppprob iprob score
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 2 HGSGTGR 2 sp|A6NLU5|VTM2B_HUMAN 0.0 0.0 1.373
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 11 RKQEEADR 3 sp|Q05682|CALD1_HUMAN 0.0 0.0 2.435
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 60 HNGTGGK 2 sp|P62937|PPIA_HUMAN 0.0 0.0 3.329
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 484 HGNSHQGEPR 3 sp|P13645|K1C10_HUMAN 1.0 0.999999 0.0001883
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 496 QMHQNAPR 2 sp|Q9NSI6|BRWD1_HUMAN 0.1448 0.00492768 1.489
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 500 IDIFQTQAEQCHIAGISQKGWNFNR 5 DECOY_sp|P47755|CAZA2_HUMAN 0.0 0.0 1.792
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 501 VQGQNLDSMLHGTGMK 3 sp|P36894|BMR1A_HUMAN 0.0 0.0 2.722
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 514 EILKIDGSNTVDHK 4 sp|Q8IZH2|XRN1_HUMAN 0.0 0.0 2.151
../mass_spectra/Adult_Adrenalgland_Gel_Elite_49_f01.mzXML 517 HGNSHQGEPR 3 sp|P13645|K1C10_HUMAN 1.0 0.999999 2.31e-05The following command line will add new data and annotation files into the existing archive.
spectroscape --add --datasearchpath /path/to/new/data Note that spectroscape will search for data (mzXML/mzML/sptxt) and annotation (.spectroscape.tsv/ipro.pep.xml/pep.xml) files recursively. Therefore new data can be organized into multiple sub-folders.
To get better control on the new data files added (e.g. excluding certain files), one can follow the command line explained in next section.
Run the following command to update the annotation of spectra in the 24 mzXML used above. One can get the interact-Adult_Adrenalgland_Gel_Elite_49.ipro.pep.xml file from a Comet+xinteract database searching pipeline in TPP.
spectroscape --run --update --updategt interact-Adult_Adrenalgland_Gel_Elite_49.ipro.pep.xmlRun the following command to add a new mzXML/mzML file.
spectroscape --run --update --updaterawdata <input>.mzXMLRun the following command can be used to search a data file. Before searching against an archive, make sure the spectral archive is annotated by search results under FDR control, e.g. annotated by pepXML files of iProphet/PeptidePropeht.
spectroscape --run --inputsource cmd --datafile <input>.mzXMLTo use the web UI, users should also download the SpectralArchiveWeb repository. If the installation is done with source
code, then SpectralArchiveWeb is already included as submodule. Here we briefly show how the web UI can be launched.
For a detailed explanation of the web UI, please refer to
the README.md file in SpectralArchiveWeb
folder.
To open the web interface, users should navigate to the SpectralArchiveWeb/scripts folder and run the following
command. The second command requires sudo.
./generate_nginx_conf.bash localhost all ../arxiv/
# the following command will require root privilege.
./start_nginx_server.bash
The command lines above will open the nginx service on local computer. Then go to the spectral archive folder, and run the following command.
# absolute path to spectroscape is required, that is why we use `which spectroscape`
# the port 8710 is currently hardcoded into the nginx configuration file, therefore do not change it.
# the only thing can be changed accordingly is the path to the SpectralArchiveWeb/arxiv/ folder.
spawn-fcgi -p 8710 -n -- `which spectroscape` --run --wwwroot ~/SpectralArchive/SpectralArchiveWeb/arxiv/After this step, we can open browser on local computer and go to the following link: http://localhost:8709. The UI will be shown as follows.
I have made two tutorial videos about how to use the web UI. Here are the links.
Wu, L., Hoque, A. & Lam, H. Spectroscape enables real-time query and visualization of a spectral archive in proteomics. Nat Commun 14, 6267 (2023). https://doi.org/10.1038/s41467-023-42006-x