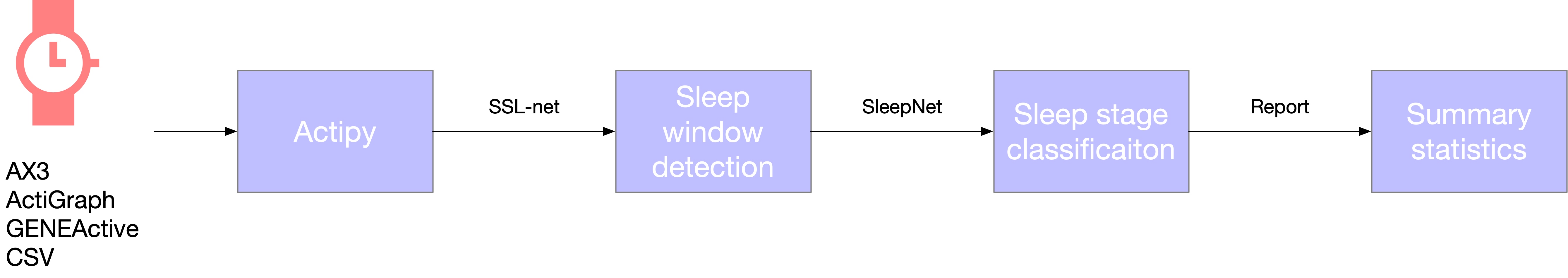
This is a Python package for classifying sleep stages from wearable sensor data / wrist - worn accelerometer. The underlying model was trained and tested in 1000 + nights of multi - centre polysomnography with tri - axial accelerometer data.
The key features of this package are as follows:
- A simple and easy - to - use API for sleep stage classification.
- Sleep / wake metric estimation including total sleep duration and sleep efficiency.
- Sleep architecture metric estimation including rapid - eye - movement(REM) / NREM sleep duration.
- Python 3.8
- Java 8 (1.8.0) or greater
Check with:
$ python --version
$ java -version$ pip install asleepAll the processing will be much faster after the first time because the model weights will to have to be downloaded the first time that the package is used.
# Process an AX3 file
$ get_sleep sample.cwa
# Or an ActiGraph file
$ get_sleep sample.gt3x
# Or a GENEActiv file
$ get_sleep sample.bin
# Or a CSV file (see data format below)
$ get_sleep sample.csvOutput
Summary
-------
{
"Filename": "sample.cwa",
"Filesize(MB)": 65.1,
"Device": "Axivity",
"DeviceID": 2278,
"ReadErrors": 0,
"SampleRate": 100.0,
"ReadOK": 1,
"StartTime": "2013-10-21 10:00:07",
"EndTime": "2013-10-28 10:00:01",
"Total sleep duration(min)": 655.7,
"Total overnight sleep(min)": 43132,
...
}
Estimated total sleep duration
---------------------
total sleep duration(min)
time
2013 - 10 - 21 435.2
2013 - 10 - 22 436.2
2013 - 10 - 23 432.2
...
Output: outputs /sample/You can visualise the sleep parameters using the following command:
$ visu_sleep PATH_TO_OUTPUT_FOLDERIf a CSV file is provided, it must have the following header: time, x, y, z.
Example:
time, x, y, z
2013 - 10 - 21 10: 00: 08.000, -0.078923, 0.396706, 0.917759
2013 - 10 - 21 10: 00: 08.010, -0.094370, 0.381479, 0.933580
2013 - 10 - 21 10: 00: 08.020, -0.094370, 0.366252, 0.901938
2013 - 10 - 21 10: 00: 08.030, -0.078923, 0.411933, 0.901938If you want to use our package for your project, please cite our paper below:
@article {Yuan2023.07.07.23292251,
author = {Hang Yuan and Tatiana Plekhanova and Rosemary Walmsley and Amy C. Reynolds and Kathleen J. Maddison and Maja Bucan and Philip Gehrman and Alex Rowlands and David W. Ray and Derrick Bennett and Joanne McVeigh and Leon Straker and Peter Eastwood and Simon D. Kyle and Aiden Doherty},
title = {Self-supervised learning of accelerometer data provides new insights for sleep and its association with mortality},
elocation-id = {2023.07.07.23292251},
year = {2023},
doi = {10.1101/2023.07.07.23292251},
publisher = {Cold Spring Harbor Laboratory Press},
URL = {https://www.medrxiv.org/content/early/2023/07/08/2023.07.07.23292251},
eprint = {https://www.medrxiv.org/content/early/2023/07/08/2023.07.07.23292251.full.pdf},
journal = {medRxiv}
}We would like to thank all our code contributors, manuscript co - authors, and research participants for their help in making this work possible. The data processing pipeline of this repository is based on the step_count package from our group. Special thanks to @chanshing for his help in developing the package.