pyCellCounter is a desktop application that can be used as an aid in the manual analysis of images taken with a microscope.
pyCellCounter is currently split into two programs. pyCellCounter can be used to manually count cells and measure distances on microscope images. Currently, only images taken with a Zeiss EM902 electron microscope and a Nikon Eclipse E800 light microscope are directly supported.
The latest release of the softwar for Windows e can be found here: https://github.com/dselcan/PyCellCounter/releases/download/0.1.0/pyCellCounter_v0.1.zip Alternatively clone the repository and add install the following dependencies: numpy, Pillow, OpenCV-Python. OpenCV can be installed by following this tutorial: http://docs.opencv.org/3.1.0/d5/de5/tutorial_py_setup_in_windows.html
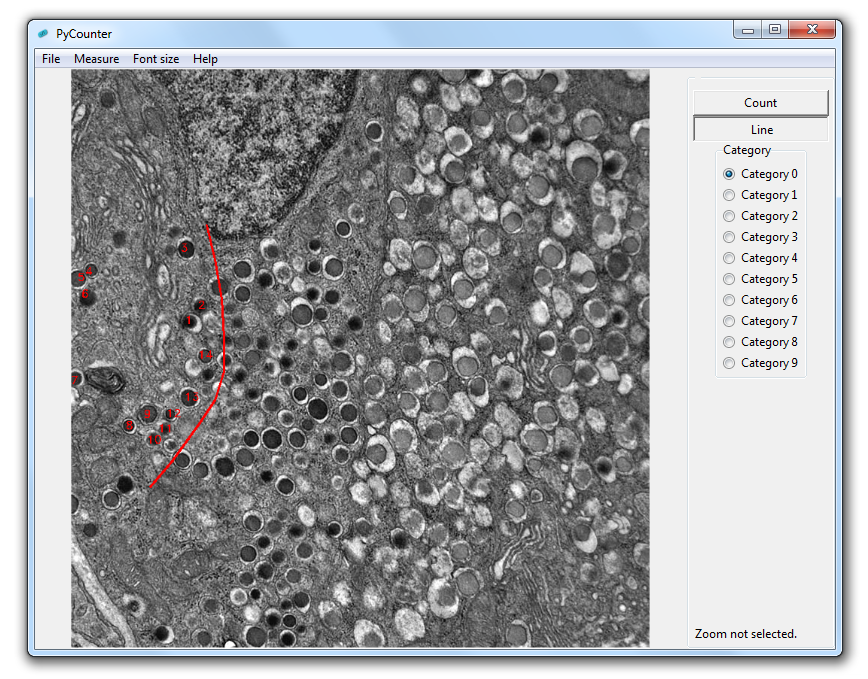
After loading an image, use the draw or line functions to count cells or draw lines. Select the zoom option to measure the distance of the line drawn. Right click to remove count points or line segments. When saving the image, the number of counted items will be appended to the bottom of the image.
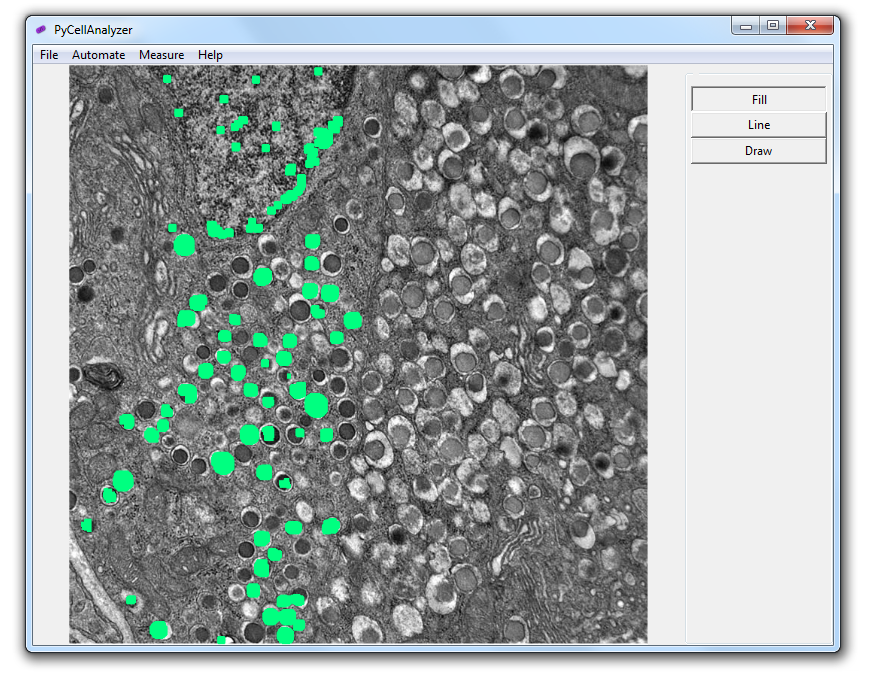
After loading an image, perform the automated analysis using the Detection menu. Afterwards, use fill (click and drag to select area), line (select lines, double click to add area) or draw (click and drag to circle the area) to add any missing cells. Right click to remove any incorrectly selected areas. When saving area data is added to the bottom of the image.
This software is licensed under a BSD license.