This repository contains a script to generate network files from any TAB-delimited observation matrix to be analyzed with Gephi.
Please let me know if you are using this script, and need more help. Contact info is at the end of this file.
An environment file is the expected input file, which consists of three columns that are separated from each other by a TAB character: unit, sample and count. Unit can be any unit (taxonomy, functions, OTU IDs, etc). Sample column contains sample names. Count column contains number of observations for a given unit in a sample. Please see the example environment file in the samples directory.
You can generate an environment file from an observation matrix with counts using the script in the scripts directory. Your input file must be a TAB-delimited matrix file. The first column should contain sample names, and first row contains unit names.
A mapping file will generate an XML file with much more information that can be used for visualization purposes from within Gephi. An example mapping file can also be found in samples directory. Briefly, the first column of the mapping file should contain all the sample names in your environment file. Any category listed in this file will appear in drop boxes in Gephi. Keyword "color" identifies a special column in this file. HTML colors given in this column color samples in Gephi directly.
To generate an example GEXF file using the sample environment file, run this from within the directory:
python generate_GEXF.py samples/oral-environment.txt
This one will take the mapping file into consideration:
python generate_GEXF.py samples/oral-environment.txt --sample-mapping samples/oral-mapping.txt
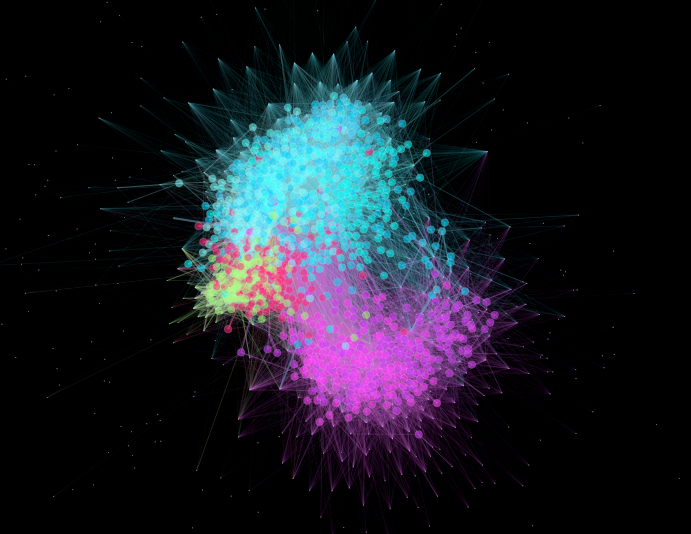
A successful run will generate samples/oral-environment.gexf, which can be opened with Gephi, and a run in Gephi with Force Atlas 2 will generate this network for the sample we just generated (it shows the distribution of samples colored by the oral sites identified in the mapping file):
If you expect more from this script, or have any questions, please let me know. You can send your e-mails to meren@mbl.edu.