nlcali is alternative method of calibrating malariasimulation runs to baseline prevalence observations by adjusting the value of starting_EIR. nlcali produces model runs using a range of EIR values and then fits a generalised linear model to the output.
nlcali is a fair bit slower than cali (run time for nlcail depends on how many simulations you can get away with running to get a good GLM fit), however it does not require setting an interval to guess EIR within, does not increase in run time as more baseline observations are added, and may be more robust to bad input since it does not require a root finder to reach convergence.
Here is some very simple code to show comparable output to cali:
# Parameters shared by both methods
set.seed(123)
# Define target, here two prevalence measures:
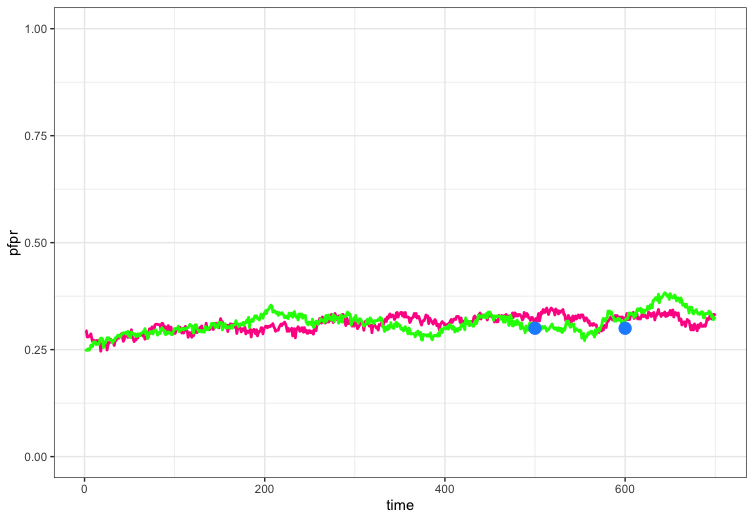
target <- c(0.3, 0.3)
# Time points at which to match target
target_tt <- c(500, 600)
parameters <- malariasimulation::get_parameters(list(human_population = 1000))
# Run nlcali, may take about 20 minutes
res <- nlcali::nlcali(parameters = parameters,
target = target,
target_tt = target_tt,
ncores = 7,
nsims = 300)
out1 <- res$stan
# Run cali
out <- cali::calibrate(parameters = parameters,
target = target,
target_tt = target_tt,
summary_function = cali::summary_pfpr_2_10,
tolerance = 0.02,
interval = c(1, 6))
# Produce a random run using the EIR value from each method
parameters <- malariasimulation::set_equilibrium(parameters, init_EIR = out$root)
raw <- malariasimulation::run_simulation(timesteps = max(target_tt) + 100, parameters = parameters)
pfpr <- summary_pfpr_2_10(raw)
parameters1 <- malariasimulation::set_equilibrium(parameters, init_EIR = median(out1$pred))
raw1 <- malariasimulation::run_simulation(timesteps = max(target_tt) + 100, parameters = parameters)
pfpr1 <- summary_pfpr_2_10(raw1)
pd <- data.frame(time = 1:(max(target_tt) + 100), pfpr = pfpr, pfpr1 = pfpr1)
ggplot() +
geom_line(data = pd, aes(x = time, y = pfpr), col = "deeppink", size = 1) +
geom_line(data = pd, aes(x = time, y= pfpr1), col = "green", size = 1) +
geom_point(aes(x = target_tt, y = target), col = "dodgerblue", size = 4) +
ylim(0, 1) +
theme_bw()