New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ICA AROMA: interacting with the other confounds #817
Comments
|
As it is right now, ICA-AROMA is run in the original BOLD time-series after: 1) Slice timing correction (if available); 2) Head motion compensation; 3) Susceptibility-derived distortion correction (if available); 4) resampling in MNI space. Your point sounds very reasonable. I have two questions at this point:
|
|
Good questions,
The obstacle to this is that ICA-AROMA depends on having an FSL warp file and affine to align the edge fraction measures with individual subjects, we can submit a pull request to ICA-AROMA to include the possibility of using the ANTs transforms, or cannibalize the ICA-AROMA code to work in FMRIPREP.
Yes, I should have thought about it more, we should also move all DVARs measures, the compcors, the region average signals, and maybe the cosine basis set, perhaps I need a primer on what the cosine basis set is. Then another question is if someone specifies |
|
Going back to the main issue I am not convinced that there is a risk of "reintroducing noise" via regression and thus a need for orthogonalization. @jdkent could you provide some evidence via simulations that this is indeed a problem? @nicholst it would be great to hear what do you think! PS This paper seems relevant: http://sci-hub.la/10.1016/j.neuroimage.2013.05.116 |
|
In the paper you've referenced, I'm suggesting the current approach is most akin to:
where we swap out bandpass filtering with ICA-AROMA, since one of the noise component selection criteria for ICA-AROMA is frequency (high frequency components are removed). If we keep nuisance regressors that may still include these high frequency components (since they were derived from the non-denoised bold) then we run the risk of re-introducing noise that ICA-AROMA originally removed. Or at least that's how I see it... the denoising process for ICA-AROMA is different from band-pass filtering so it may not have an impact. |
|
I understand your reasoning, but the assumption is that regression and bandpass behave the same way. Here's a though experiment that makes me skeptical. Lets say I have a variable A from which I regress out variable B (fit a model to get residuals). This gives me variable C which has no shared variance with variable B (by definition). Now I will do the same thing again - regress variable B from variable C. Would this "reintroduce" variance from variable B that was previously removed? I doubt it, but we should simulate this. |
|
Chris & co,
I’m just finalising a blog post to flesh out my response to our conversation over the summer stimulated by your “What is aggressive/non-agressive?” python notebook. Here is what I would say is the relevant take away:
Consider response (BOLD data) Y, some signal of interest X and confound C.
In an ideal world we’d fit everything together, at once:
Y ~ X + C
in R-style linear modeling notation, giving inference on effect of X while controlling for C. Let’s call this the ‘gold standard’, the result we want to replicate.
In practice, we want to provide adjusted data for diverse uses, so we can instead do it in stages in one of two ways and get the very same inference.
Approach 1.
Fit the only the nuisance
Y ~ C
making what we can call Ya1, adjusted data. With this, if we want to get the same inferences as the gold standard then we must run the following regression:
Ya1 ~ Xa
where Xa comes from the adjustment of X, residuals from `X ~ C`. Note, an equivalent approach is to just leave X as is and include C in the design...
Ya1 ~ X + C
… Ya1 doesn’t need the C, but adding C to the regression automatically implements the C-adjustment-of-X.
Approach 2.
Alternatively, if X is available when we are doing preprocessing, we could fit the gold standard model but *only* subtract off the estimated nuisance effect of C (this corresponds to the “non-agressive” setting, where C is all bad ICA, components, X all non-bad ICA components, and only the C*beta portion of the fitted model is subtracted off). Call the adjusted data Ya2.
In this case, perhaps surprisingly, you can do subsequent analyses ignorant of C
Ya2 ~ X
and you’ll get the gold standard inference.
So… if we are intent in obtaining what would be the gold standard analysis we have two choices: (1) Have the final actual regressors of interest available at the time of preprocessing, or (2) Keep the nuisance regressors around, available at the time of final inference.
Make sense?
PS: In all of these settings, to get the DF right you’ll have to keep track of the the previous regressions.
|
|
@chrisfilo, here is the simulation code. Just used the code you provided from your ICA_AROMA investigations. In the simple case you provided, your intuition was correct, the same regressor applied twice does not result in different outcomes. My intuition wasn't correct, I'll read more about the differences between bandpass filtering and regression. I don't know if I need to model any additional complexities to make the simulation more valid. Thanks @nicholst! I'll need to digest that for a bit. The only comment I'll add is that we are additionally interested in including confounds that were derived from other methods (e.g. framewise displacement, DVARS) and if it's necessary to "denoise" the other (non ica-aroma) confounds so they don't add variance that ICA_AROMA removed (as in the paper @chrisfilo referenced), but I'm starting to think this was an ill-formed question. |
|
@chrisfilo I took another stab at testing ica_aroma between extracting confounds before or after denoising, and found different results depending on whether the confound was extracted before or after denoising, please see and check my work |
|
Since global signal is a spatial average I don't think it would work the way you expect. In non-agressive denoising each voxel is denoised in different way so averaging all of them (when calculating global signal) will yield a different regressor. I don't think it is that surprising that you are getting a different result depending on the order. @nicholst thanks for the explanation. Indeed we are dealing with a slightly different question here: if you do non-aggressive denoising to the data and you later want to regress other sources of noise (say motion) would that additional regression reintroduce noise previously removed by AROMA? I don't think it would. |
|
thanks @chrisfilo. Then if it's not surprising the results are different, should it be necessary to keep the global signal as close to the data as possible (as well as other measures that use multiple voxels to inform their values, such as acompcor & tcompcor)? That is, calculate the confounds from the denoised data as is proposed in #959? (assuming people are interested in the non-aggressive denoising). |
|
I don't have a strong opinion on this. I don't think either approach is wrong. Might be worth searching the literature to see if someone did compare those strategies. |
|
Do they say which stage are nuisance regressors estimated at?
Best,
Chris
…On Mon, Feb 5, 2018 at 4:54 PM, James Kent ***@***.***> wrote:
I couldn't find a comparison between the two approaches, but the Pruim
(2015) paper does the nuisance regression after denoising of the bold
series.
[image: image]
<https://camo.githubusercontent.com/2d4625ac43953c1654ac305da117806a09b4f331/68747470733a2f2f6172732e656c732d63646e2e636f6d2f636f6e74656e742f696d6167652f312d73322e302d53313035333831313931353030313832322d6772312e6a7067>
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#817 (comment)>,
or mute the thread
<https://github.com/notifications/unsubscribe-auth/AAOkp4gjdVfJW9Uk72df9FX7PhrHWk3Tks5tR6LHgaJpZM4QVx-A>
.
|
|
page 269: first paragraph under methods/ICA-AROMA
I'm assuming that means the nuisance confounds were extracted and applied at that time, perhaps a bad assumption? |
|
Probably correct. I see your point. The only problem now is that if we estimate global signal, CSF, WM etc. after applying non-aggressive denoising those regressors will only be usable with the denoised data. Should we even save non denoised data then ICA AROMA is requested? Such situation may lead to a risk of users using wrong regressors. Alternative is to include a WDYT @oesteban? |
No, that isn't possible. Non-agressive denoising isn't the GLM, and so there certainly can be variance left in the data that can be later be removed by other methods (even the very temporal ICA components used in non-agressive denoising). Each successive "agressive" correction and only reduce the total variance in the data. Update: 2 Oct 2019 - As pointed out by @mangstad, this reply is wrong/misleading. While it is true that subsequent nuisance regressions can only reduce the total sums-of-squares, due to correlation among nuisance variables, subsequent nuisance regressions can in fact re-introduce variance related to a previously-regressed-out nuisance variable. The solution is to do all nuisance regression at once as part of a multiple linear regression, or use careful orthogonalisation; see https://www.ncbi.nlm.nih.gov/pubmed/30666750 for more. |
|
Thank you for your insight @nicholst. Perhaps I am missing something here, I've made another toy example to try to demonstrate the issue. Let's say we derived nuisance confounds from the bold data before denoising with ICA-AROMA, and then do non-aggressive denoising. If I then wanted to regress the nuisance confound from the denoised bold data (e.g. P.S. Thank you all for this stimulating conversation! |
|
Hi @jdkent, Nice example, but my point was solely about total sum-of-squares: Residuals will always be smaller with any additional regressor. I'm not yet a Python head... in Matlabese Y=[1:100]';
X=[ones(100,1) rand(100,1)];
betahat=pinv(X)*Y;
Yhat=X*betahat;
plot(Y,Y-mean(Y),Y,Y-Yhat)
[mean((Y-mean(Y)).^2) mean((Y-Yhat).^2)]
% I got : 833.2500 811.2734But, certainly, there can be variation in the regressor that doesn't help (as is visually seen with the 'jaggies' in your example) but to the extent that there is any overlap between the nuisance and the data it will remove something. Does this help? |
|
Yes, that helps tremendously @nicholst, thank you! Now I see what you are referring to. With that in mind, I see my concern is not really with increasing variance (which is impossible), it's about adding structured noise to the residual bold series (This is one of those epiphany moments 😄). The structured noise could come from breathing/heart rate/other physiological factors, movement, and anything else ICA-AROMA happens to classify as noise. If the nuisance confounds were derived from the non-denoised bold, the worry is that this structured noise may be added back in to the denoised bold residual (represented by the jaggies in the example). This would be bad for any functional connectivity analyses as this may introduce non-neurally based correlations between brain regions. Thank you both @nicholst @chrisfilo for putting up with my incorrect use of terms; I think I've finally caught up. |
|
Just to clarify, the 'jaggies' are reflections of living in the 'orthogonal subspace' of the nuisance… i.e. residuals must be orthogonal to the regressor space, and hence will reflect their 'complimentary' structure. In general if the nuisance is completely irrelevant (unrelated to anything) such adjustment shouldn’t worsen any subsequent analysis. But if the nuisance *is* related to the signal of interest at all then it will be important to equivalently adjust the signal of interest.
|
|
Thank you. Your follow-up inspired me to run some more tests in this notebook to get an intuition of what would happen if I took two timeseries (y1 and y2; or voxel1 and voxel2), and regressed the same nuisance confound (representing the confound pulled from non-denoised BOLD data) from each of them. For the outcome measure, I compared the correlations between the residuals and between the original Sometimes the correlation increases, and I think this is due to how the pearson's correlation coefficient is calculated since it includes the variance of y1 and y2 in the denominator. So in the scenario where the variance decreases in y1 and y2 more than the decrease of covariance between y1 and y2, the pearson's correlation would be inflated. So while I think the covariance between But this is kind of a divergence from my original concern, and if anything it makes it harder for me predict whether a regressor derived from the non-denoised bold signal is worse than a regressor derived from the denoised bold signal. I will think further upon this, and thanks again! I've now effectively buried @chrisfilo's question to @oesteban, sorry:
We should probably move this conversation back to the pull request, now that we are dealing with the code implementation of the solution #959. |
|
Hi, I am pretty swamped right now to process all of this but will do soon. Thanks so very much you all for this great conversation! I just wanted to make sure we don't miss a bit of detail here. For that, I would suggest both @chrisfilo and @jdkent to add their notebooks into the fmriprep repo. We could have a It could be done in separate PRs, as it is just documentation. Alternatively/additionally, we could have them rendered to rst and added in the documentation. WDYT? As regards where to keep the conversation, it is probably best to keep it here since all the details are given above. EDIT: I'm also thinking on adapting @nicholst's comments to our documentation. |
|
@jdkent I know it's not as appealing as simulation, but just to confirm in math mode: If Y_1 and Y_2 are independent, no transformation done to each individually can induce dependence: If we have two time series Y_1 and Y_2 that are independent (but possibly have their own autocorrelation) |
|
Math mode is definitely appealing, I'm just not well versed, but working on it, albeit slowly. 😄 @oesteban: I'll work on making a more sensible story with the code bits worked up, most of it is me not clearly understanding the problem, and then using incorrect terms to describe the problem. Furthermore, I am no longer convinced that it is definitely a problem, but maybe a problem. Hopefully will make more sense in the write-up. |
|
Hi all, Thank you for all your posts here. I learned a lot!
Thanks! |
|
I'm not sure how you can do that with FMRIPREP. One thing is certain ICA AROMA depends on your data being in the MNI space (or being able to put it in the MNI space) because it uses ROIs defined in MNI. Out of curiosity which template are you using as coregistration target for your infant study? |
|
oh! I thought that this thread is in the ICA-AROMA git. It is my bad; i did massive google search, so I lost my way. ;p I am using ICA-AROMA out of FMRIPREP. You are right that ICA-AROMA plays on the MNI space by converting the native image to MNI template. However my understanding is that ICA-AROMA run Melodic on the native space first, and then normalize only melodic_IC.nii.gz to MNI space (melodic_ic_thr_MNI2mm), using reg matrix I put in the command line, to match noise property they have based on 2mm MNI space. Therefore I guess it would be okay to regress out classified-noise ICs for the native image since melodic_mix file, ICA-AROMA uses for denoising, still is from the native image. it is not optimal for spatial properties though. But not sure. For baby template, I am using UNC's IBIS baby template (https://www.med.unc.edu/bric/ideagroup/free-softwares/unc-infant-0-1-2-atlases) |
|
Hi, I'll try to close this discussion with permission from @chrisfilo, @jdkent and @nicholst. After re-reading the whole thread, my impression is that we are safe to keep FMRIPREP working as it is implemented now, and recommend the following: If you are to fit the "aggressively" denoised time-series, you can still use other regressors (with the exception of global signals?) if you orthogonalize them against the "non-aggressive" confounds.
Thanks! |
non-aggressive case: Based on the notebook, my current opinion is that one should be able to the use other confounds (such as global signal) without modification even if they were derived from the non-denoised data. aggressive case: also should be okay to use the global signal without modification as well since you will just be including the noise columns in the confounds |
|
Thanks very much. I'll go ahead and close this issue. Please reopen if I misinterpreted your message. Please close #959. If I'm not wrong, that one should not be necessary anymore. Is that correct? |
|
I have proform preprocessing by fmriprep. And there are some files created, like I have some questions as follow:
|
|
Hi @yuan0821. Closed issues are easy to skip over, so in general, it would be better to open a new issue, and link to this one, for example: But while we're here:
|
|
Got it! just opened an issue (#1804) so I can keep track of it. |

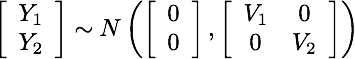

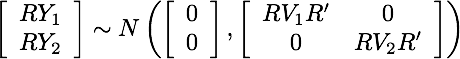
The standard way to use ICA-AROMA with non-aggressive denoising in conjunction with other confounds in the tsv (specifically acompcor, tcompcor, and other measures derived from the bold), is to pull out those signals after denoising of the data, as to not re-introduce noise removed by ICA-AROMA (if we calculated acompcor, tcompcor before ICA-denoising). So my question is: does this mean we should re-organize the code to also pass in the non-aggressively denoised bold to calculate these other measures? or would it be equivalent to do full regression removing the variance explained by the "noise components" on the other confounds in the tsv as I did in the final part of this jupyter notebook. The latter would be substantially easier, but I don't know if we are sacrificing something, or doing something not recommended.
The text was updated successfully, but these errors were encountered: