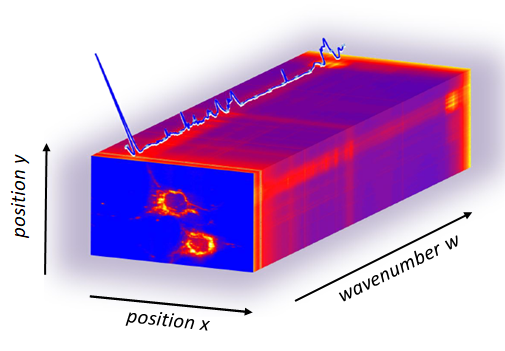
A Raman hyperspectral image consists of a three dimensional matrice, the first two axis corresponding to the pixel position and the third axis being the Raman intensity spectrum at that pixel. One image contains roughly 80000 spectra with 1000 wavenumbers each, in the range of 700 - 3000 cm-1. In addition to experimental artifact (laser focus, sample preparation), Raman data typically involve significant noises, fluorescence background due to water and substrate, as well alteration by cosmic rays. This repository contains the tool to process Hyperspectral Raman images and make it as free as possible from differences in experimental conditions:
Process the hyperspectral Raman image with:
- Cosmic ray detection
- Correction for Irradiance profile
- Identification and subtraction of Background spectrum
- Singular Value Decomposition Denoising
- Polyfit fluorescence correction
- Spectra area normalization
- Cell region identification
- Nucleus and Cytoplasm spectra differentiation
To process Raman images, run src/_Main - Raman_analysis.py. Running the program requires python3, and in addition to standard libraries such as numpy or matplotlib, the program also requires hdf5storage to read .mat files, and brokenaxis to plot the spectra. Two raw images are provided in src/Raw_Measurements.py to show how the code works, but more Raman images taken on two different devices are publicly available [Device 1, Device 2]
[1] Pelissier A, Mochizuki K, Kumamoto Y, Taylor JN, Clement JE, Fujita K, Harada Y and Komatsuzaki T. Raman Diagnosis of Thyroid Carcinoma in Co-cultured System with Minimizing Extrinsic Background across Different Devices. arXiv. 2023
[2] Taylor JN, Pélissier A, Mochizuki K, Hashimoto K, Kumamoto Y, Harada Y, Fujita K, Bocklitz T, Komatsuzaki T. Correction for Extrinsic Background in Raman Hyperspectral Images. Analytical Chemistry. 2023 Aug 10;95(33):12298-305.