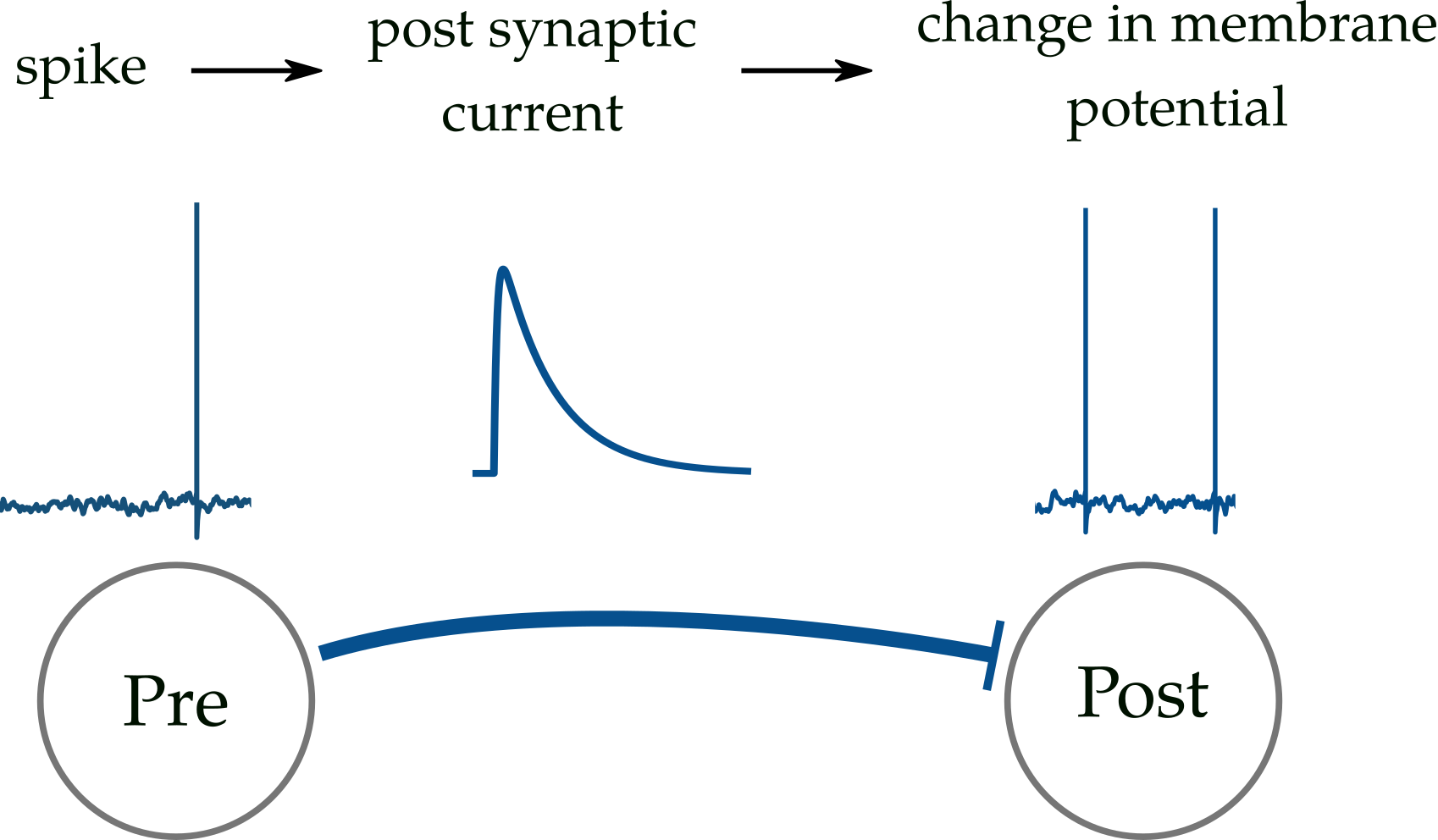
This repository aims at introducing a spiking neural network simulator along with providing instructions on how to implement such networks. With this simulator, one can build leaky integrate and fire (LIF) neural networks with realistic synaptic transmissions and desired neural structures and inputs.
This is a detailed biological model in which each neuron is represented by a variable (membrane potential). When the membrane potential of a pre-synaptic neuron crosses a threshold, it emits a spike that consequently creates a post-synaptic current that changes the post-synaptic mebrane potential. This is why it is called leaky integrate and fire, as the membrane potetial decays (leaks) to a rest potential, integrates incoming inputs and fires when it crosses the threshold.
The simulator basically runs a giant dynamical system, with the dynamical variables representing membrane potentials of neurons. Generally, one can use any sort of connectivity matrix in this simulator. Additionally, there are built-in functions that can construct multi-population networks by creating the appropriate connectivity matrices with given statistics. Moreover, any time-dependent input can be defined to feed into the network. Built-in functions are designed to construct random Poisson inputs with given statistics similar to those of cortex.
The simulator outputs the selected neurons membrane potentials and spike times. Using these outputs, raster plots and other visuals can be plotted using incorporated functions. Several statistical measures can also be calculated by built-in functions.
To see how this simulator is used in a research project, please refer to the network simulation part of our publication:
Non-monotonic effects of GABAergic synaptic inputs on neuronal firing
A Abed Zadeh, BD Turner, N Calakos, N Brunel
PLOS Computational Biology 18 (6), 2022
This simulator consists of three libraries as follows:
- spiking.py: This library is the main module for dynamics of a neural network. It consists of codes for initializing a network, running the dynamics on that network, visualizing the output, and performing statistical analysis on the dynamics.
- populations.py: This library consists of predefined populations to be used for neurosimulator. There exist single population and multi-population networks of striatum with inferred connectivity profiles from experimental literature.
- plasticity.py: Here online plasticity rules can be defined to be added to the network dynamics. A predefined anti-Hebbian rule is incorporated. With an online plasticity rule, connectivity weights of the network change depending on neural network activity such as pre-post neural firing rates.
a network can be defined and run using few lines of code as simple as below:
import neurosimulator.spiking as ns
from neurosimulator.populations import Striatum
ns.initNetwork(Striatum)
(SpikeTime, Spike, V) = rundynamics(endtime=1000, monitor_ind=[0,1,2,3])The above code imports neurosimulator and from populations the striatum predefined population, consisting of three different cell types (FSI, dSPN, iSPN). Then it initializes the network in the simulator using initNetwork(). Using rundynamics() the simulator runs the network for duration of endtime in ms and monitors the selected neurons (with indices monitor_ind) and outputs their spike times (SpikeTime), binary spike arrays in time (Spike), and membrane potentials (V).
To visualize the dynamics of the network, there are several visualization functions in neurosimulator. A built-in function can run and visulalize the dynamics at the same time using:
ns.runplot(1000, ns.monitorind([10,10,10]));Here, using runplot(), the simulator runs the network for 1000 ms, chooses 10 randomly selected neurons from each three populations by monitorind([10,10,10]) and plots the rasterplot of spike activity and average population activity across time.
One can turn on or off placticity rules at any stage of simulation. Here is a sample code on how plasticity can be incorporated in the model
import neurosimulator.spiking as ns
from populations import Striatum
ns.initNetwork(Striatum)
ns.runplot(1000, ns.monitorind([10,10,10]));
from neurosimulator.plasticity import AntiHebbian
ns.initPlasticity(AntiHebbian)
ns.runplot(1000, ns.monitorind([10,10,10]));
from neurosimulator.plasticity import Off
ns.initPlasticity(Off)With this code the striatal network runs for 1000 ms and then anti-Hebbian plasticity is turned on and the network runs for another 1000 ms and then plasticity is turned off.