Pointers to publicly available assembly, binning, and profiling results (with structured metadata).
-
Fork (i.e. make a copy of) the data repository to your GitHub account here.
-
Get a local copy of the forked repository on your computer:
git clone https://github.com/<USERNAME>/data.git- To be able to pull changes from the original repository, add the original data repo as an upstream remote:
cd data
git remote add upstream https://github.com/CAMI-challenge/data.git- Make sure that the master branch is up to date with the original data repository:
git checkout master
git pull upstream master
git push origin master- Create and checkout a new branch for your work:
git checkout -b update_metadata- Enter the matadata in the appropriate tab-separated table, e.g. in CAMI2/toy/mouse_gut/taxonomic_profiling.tsv. As your results will still be uploaded and do not yet have a DOI, enter the word
newin the DOI column.
You may also create new directories and tsv files. Every directory with tsv files must contain adescription.txtwith the title of the respective CAMI data set. The upload tool will scan every subdirectory in the data repository for tsv files listed in tasks.tsv.
Notes about the table columns:
In column SamplesUsed, provide the numbers identifying the samples used. If multiple samples were used, you can provide ranges, e.g. 0-63, and specific samples, e.g. 0-63,70,75.
Multiple files can be provided in the FileName column by separating them with semi-colons, e.g. file1;file2;file3.
Similarly, multiple creators, ORCIDs, and affiliations can be provided with semi-colons in columns Creator, ORCID, and Affiliation.
-
In your Zenodo account settings, go to Applications and create a personal access token with
deposit:writepermission. You may also activate thedeposit:actionspermission, or leave it deactivated to avoid accidentally publishing results. Once the token is created, store it safely! -
Upload your files using the upload tool (requires Python 3):
./zenodo_upload.py -h
usage: zenodo_upload.py [-h] [--github_dir GITHUB_DIR] --files_dir FILES_DIR
--zenodo_token ZENODO_TOKEN [--sandbox]
CAMI Zenodo upload tool
optional arguments:
-h, --help show this help message and exit
--github_dir GITHUB_DIR
GitHub directory [default: current working directory]
--files_dir FILES_DIR
Directory containing files to be uploaded
--zenodo_token ZENODO_TOKEN
Zenodo access tokenExample:
./zenodo_upload.py --files_dir /home/me/myresults/ --zenodo_token xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx--files_dir is the path to the files of results that are being uploaded. It should be different from --github_dir, as those files are not supposed to be uploaded to GitHub.
--zenodo_token is the token that you generated, as described above.
WARNING: ONCE YOUR RESULTS ARE PUBLISHED, THEY CANNOT BE DELETED!
-
To publish your results, you can either go to your list of uploads on the Zenodo website, select an upload, make final changes if needed, and then publish it by clicking on the publish button.
-
Alternatively, you can use the publish tool (remember to activate the option
deposit:actionsfor your token on Zenodo):
./zenodo_publish.py -h
usage: zenodo_publish.py [-h] --zenodo_token ZENODO_TOKEN [--sandbox] logfile
CAMI Zenodo publish tool
positional arguments:
logfile Log file containing deposition IDs
optional arguments:
-h, --help show this help message and exit
--zenodo_token ZENODO_TOKEN
Zenodo access token
--sandbox Sandbox testThe logfile, called zenodo_deposits_DATE_TIME.log is automatically created by the upload tool in the working directory, where DATE_TIME are the date and time of the upload.
-
Update the metadata tables by replacing the word
newwith the generated DOIs. -
Choose the edited files to commit, e.g. data/CAMI2/toy/mouse_gut/taxonomic_profiling.tsv:
git add data/CAMI2/toy/mouse_gut/taxonomic_profiling.tsv- Commit the changes with a sensible message:
git commit -m "add results for tool x"- Push your commit to GitHub:
git push- Create a pull request on GitHub for your forked data repository to the original repository (see Creating a pull request from a fork).
Submit your results to zenodo's CAMI Community - https://zenodo.org/communities/cami -, click on New upload.
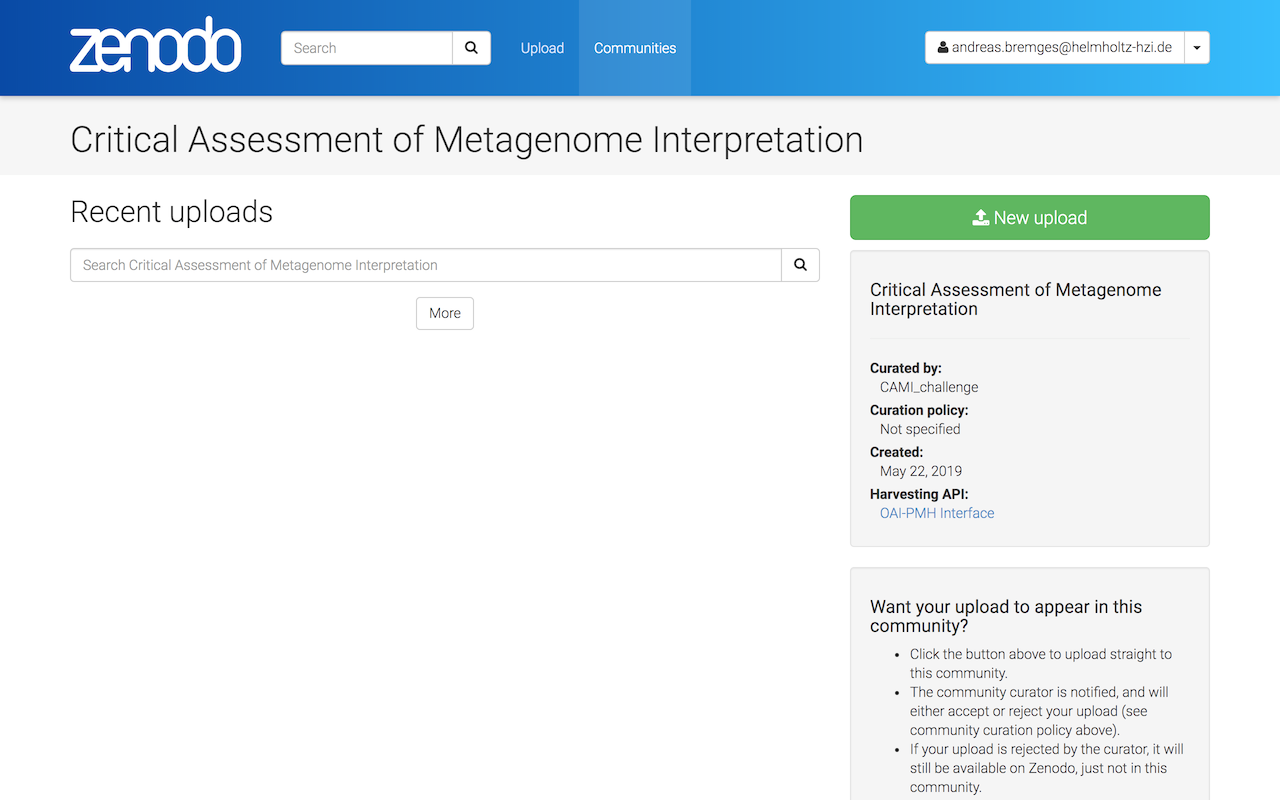
Drag and drop your result files or Choose files.
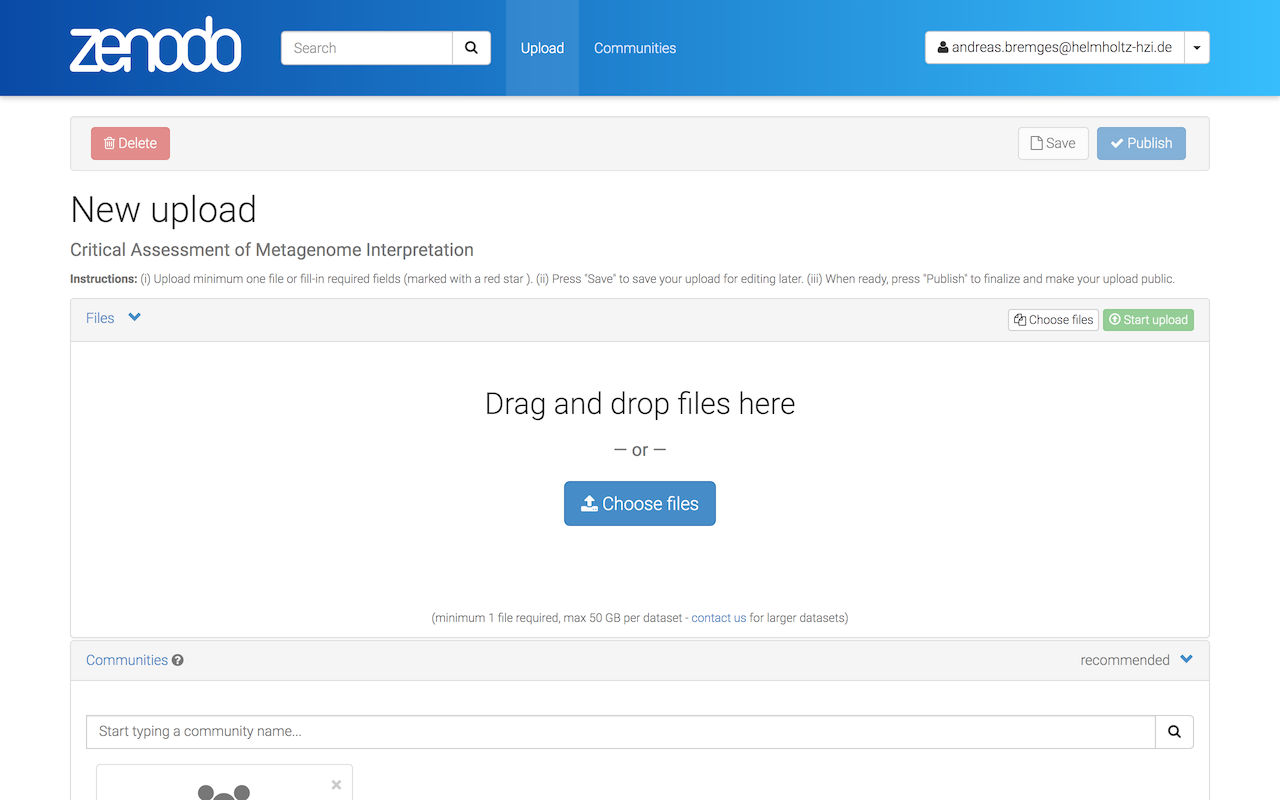
Click on Start upload (this might take a while).
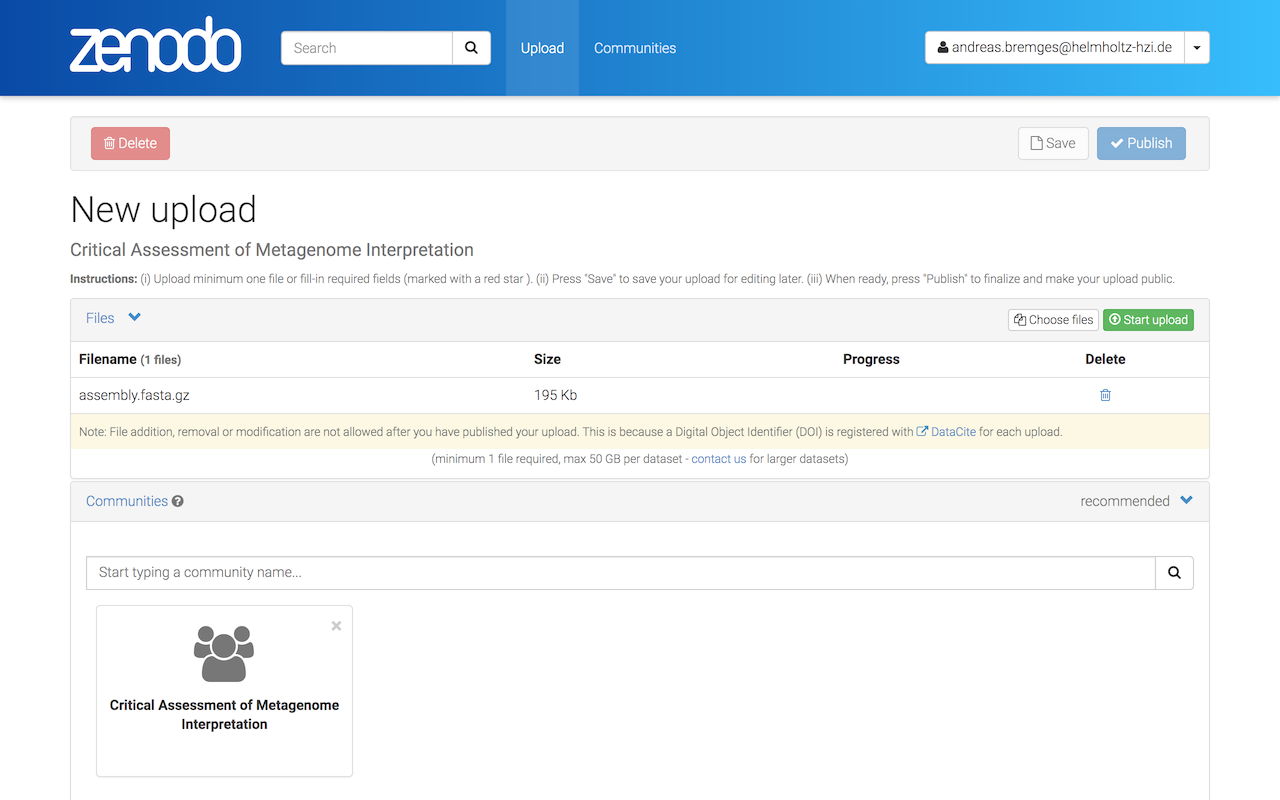
A checkmark (under Progress) indicates that the upload was successful.
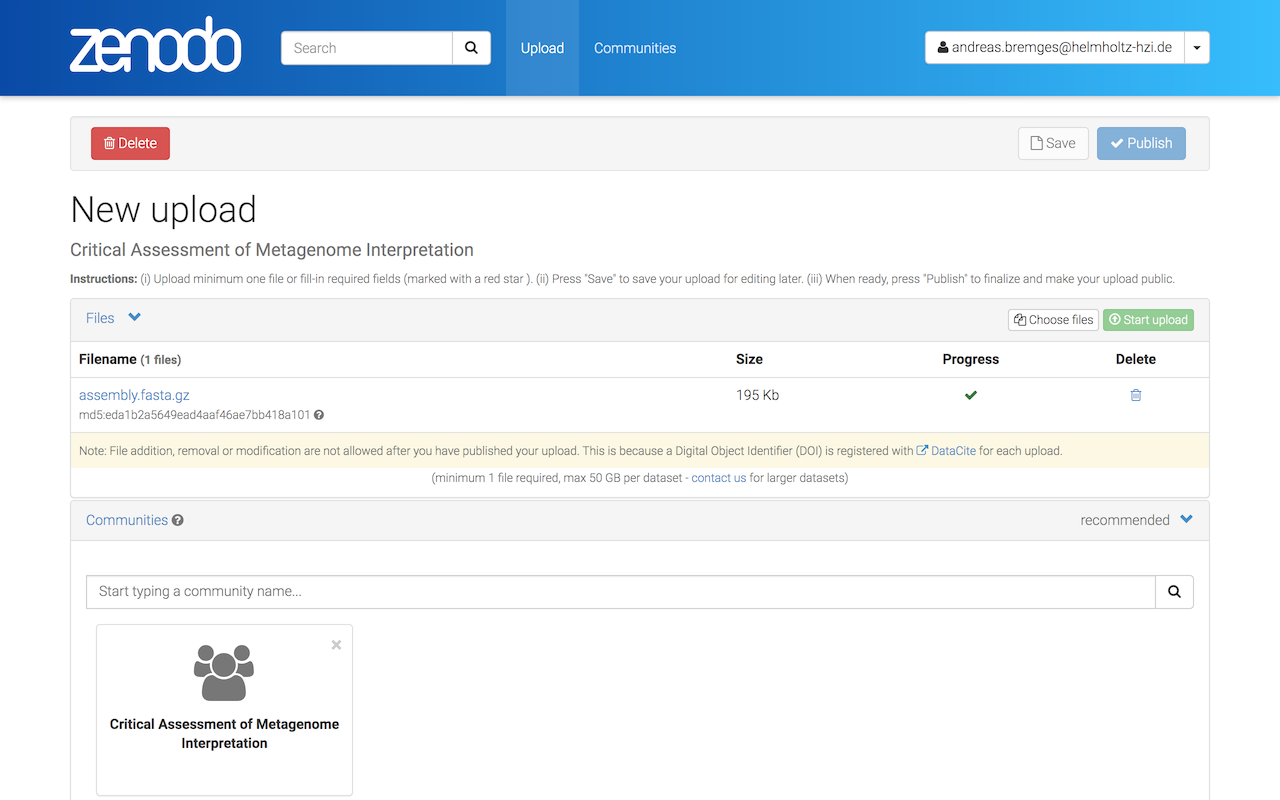
As the Upload type, select Dataset and enter Title, Authors, and brief Description (you will provide structured metadata in Step 2).
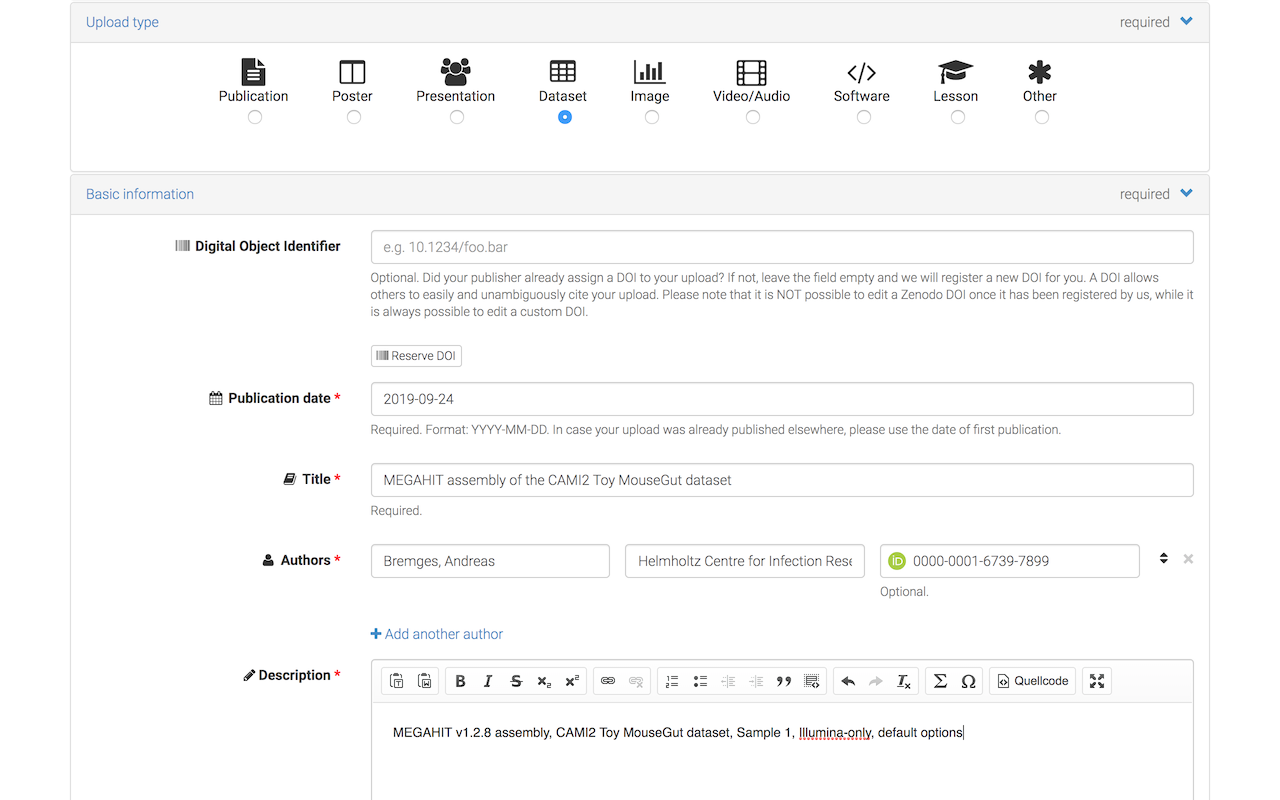
Select Open Access (or Restricted Access, if you insist on keeping your submission private).
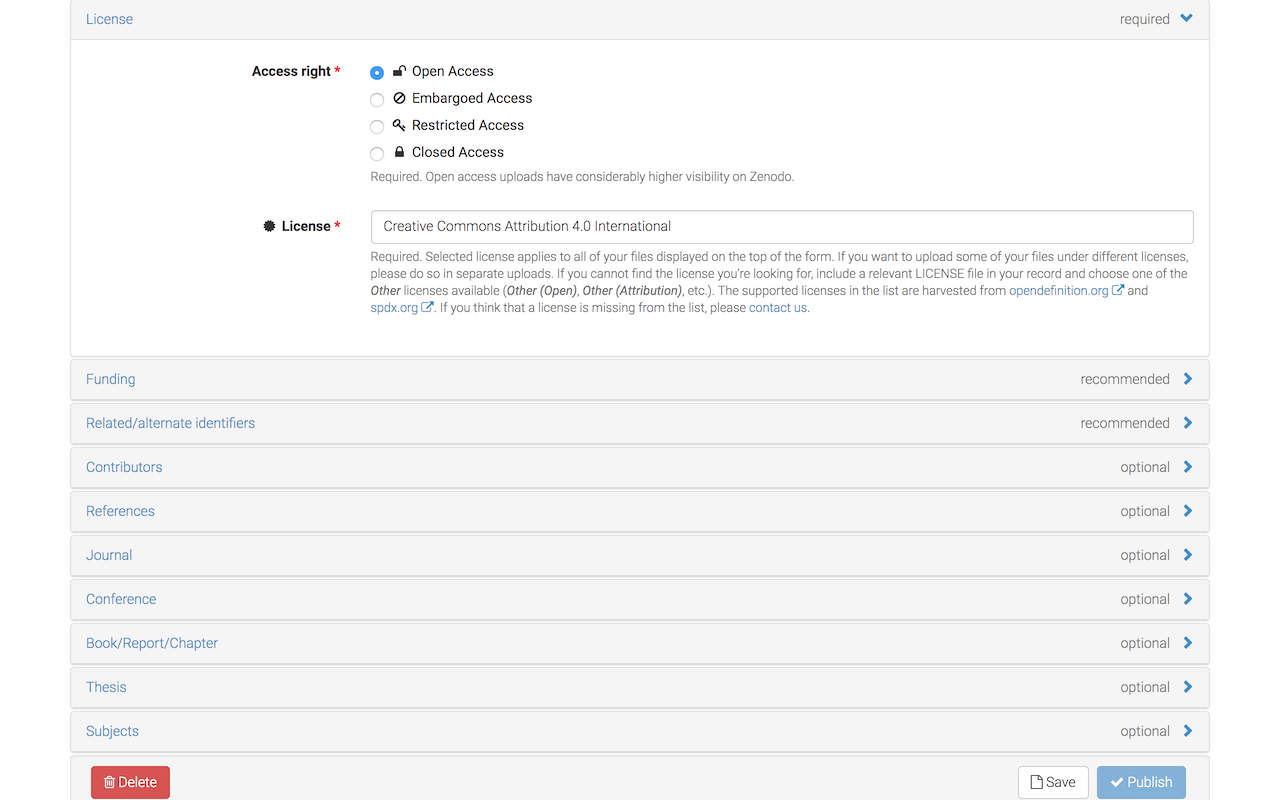
Click on Save and carefully review your submission files and metadata.
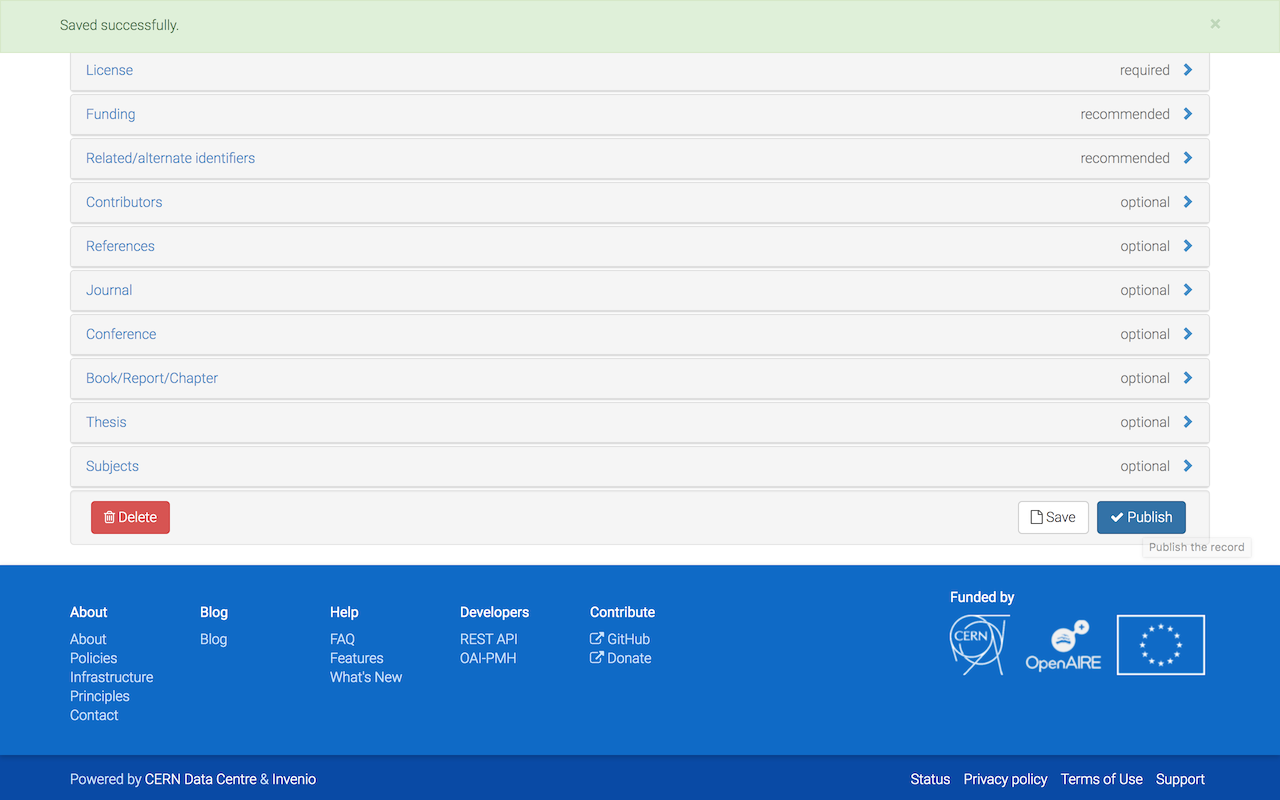
Click on Publish and confirm that you want to assign a DOI to your submission.
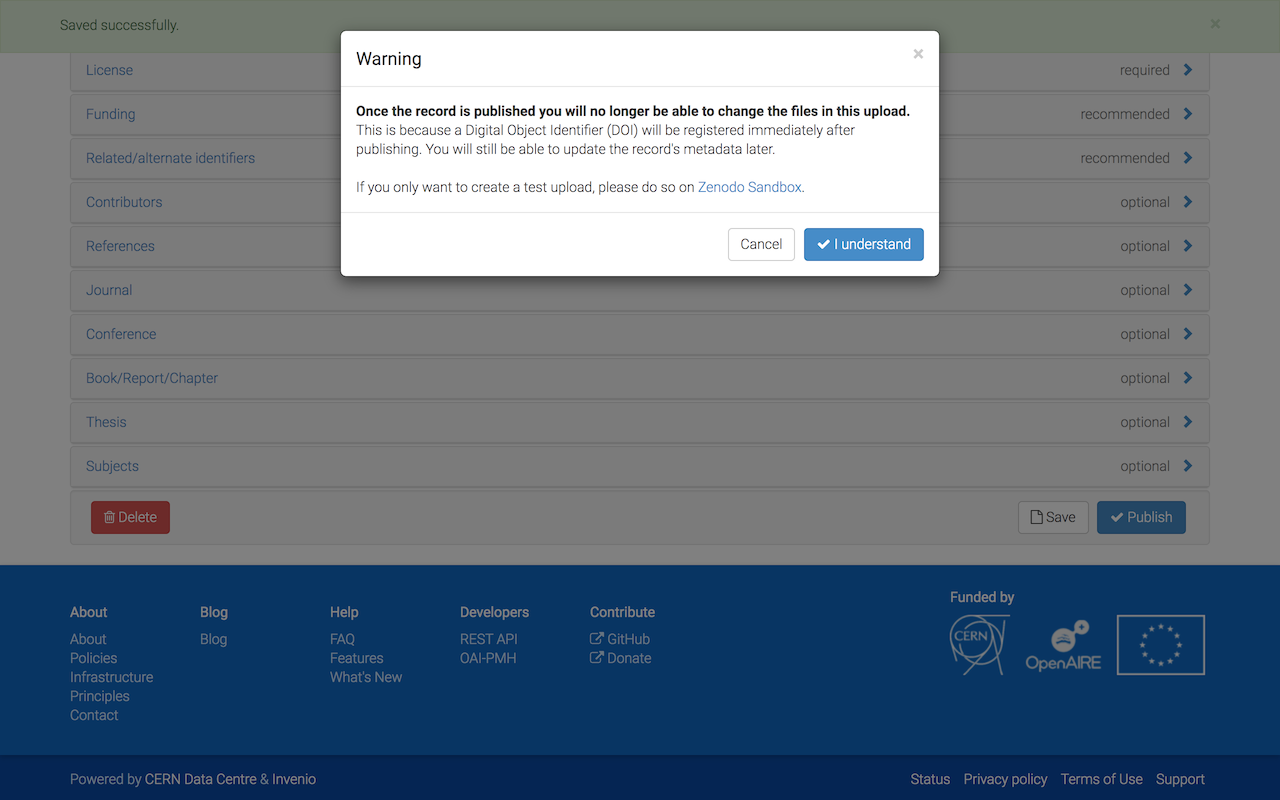
Congratulations, you successfully uploaded your results to zenodo!
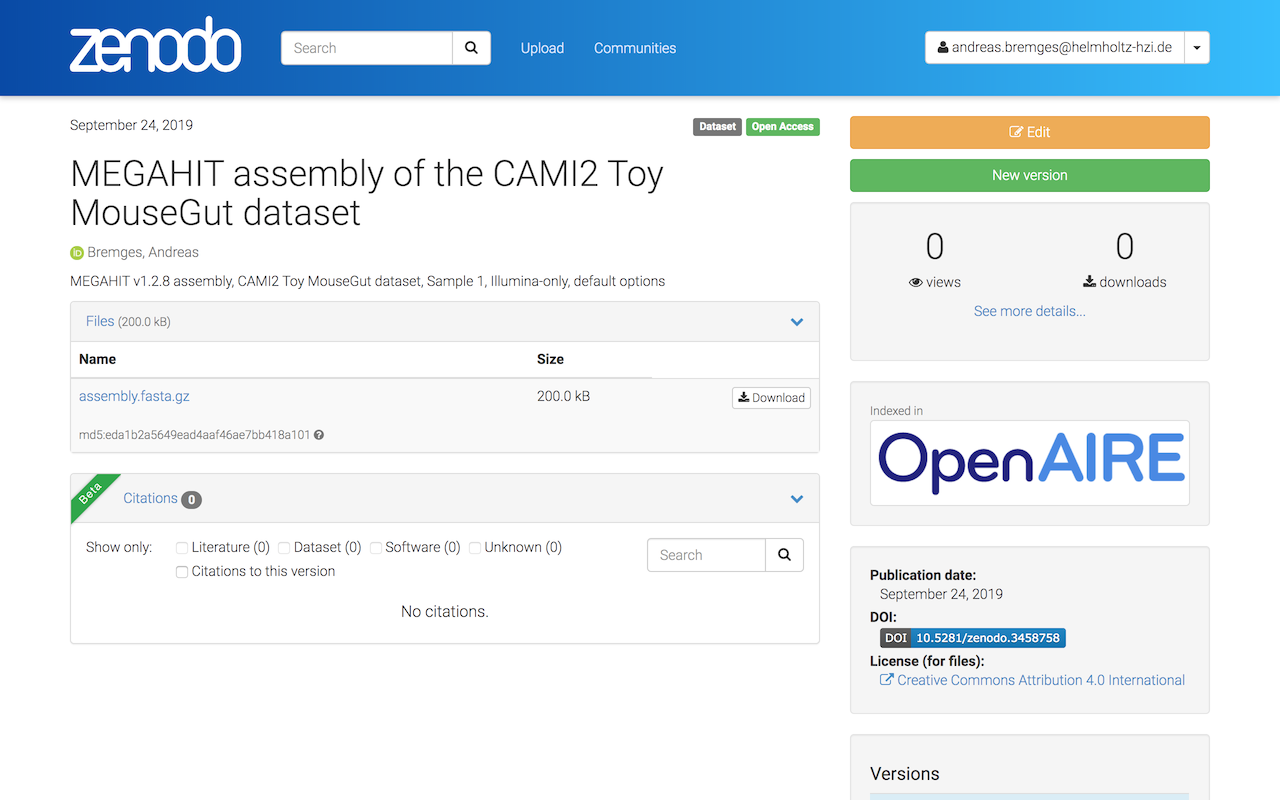
We will review your submission (after you completed Step 2) and upon approval the CAMI Community badge will appear next to your submission.
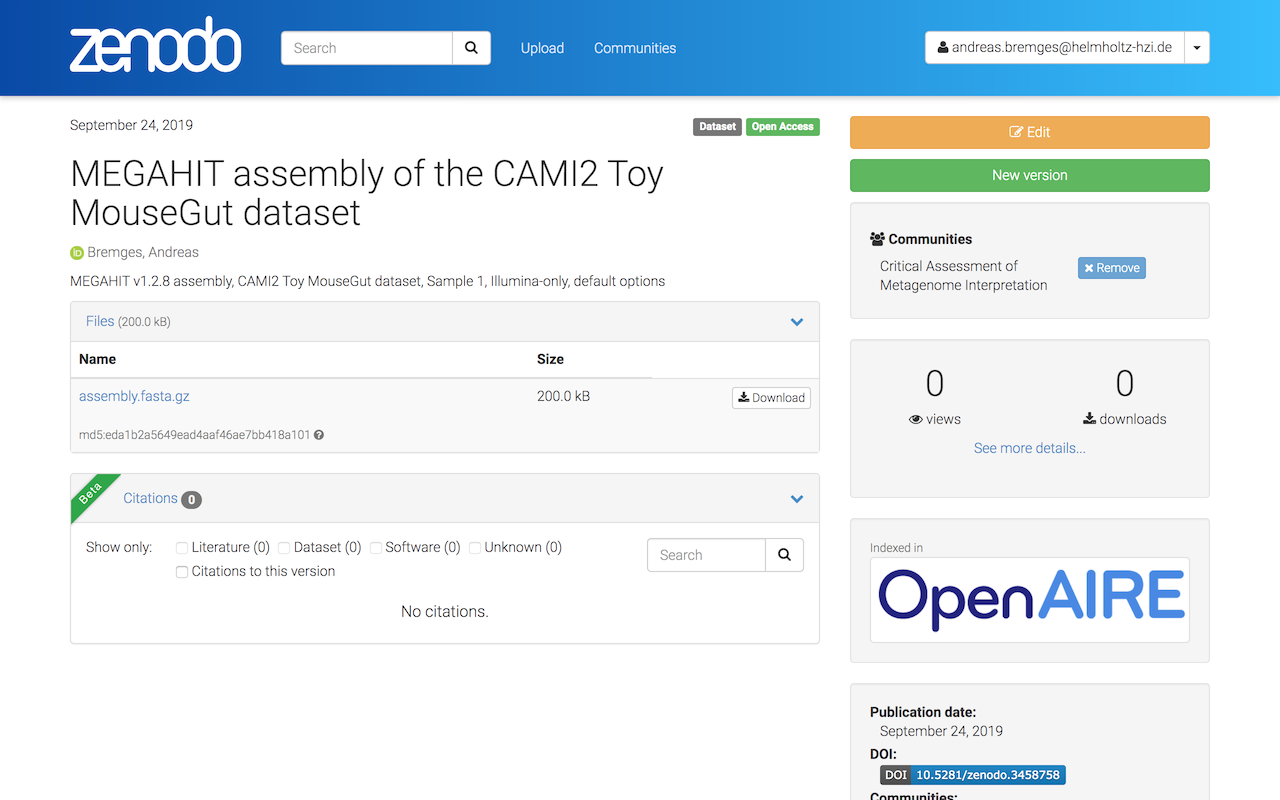
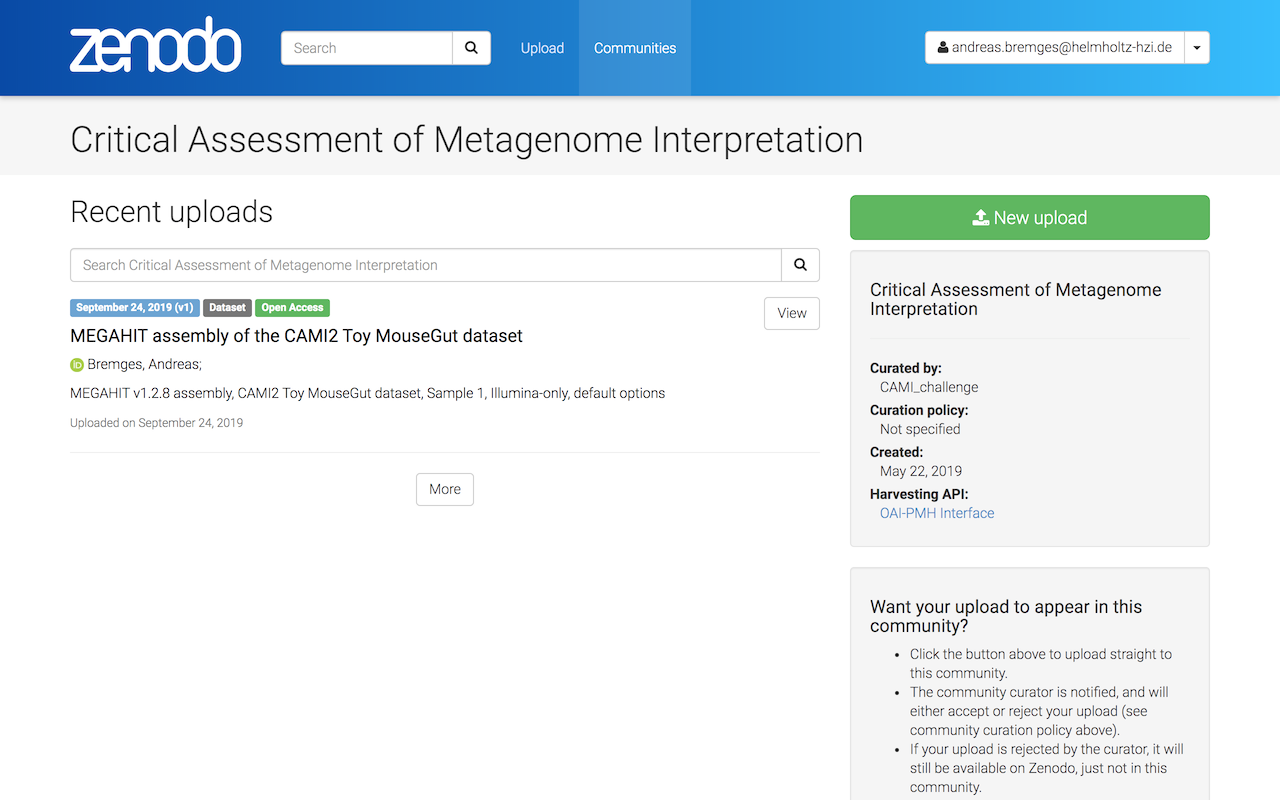
Additionally, your submission will be visible also on the CAMI Community site.
Metadata and links to the raw data are stored in this repository in tab-separated files, one table per dataset (e.g. CAMI2/Toy/MouseGut/assembly.tsv).
Please add your metadata by following the GitHub flow: clone the repository, add one or more row(s) to the appropriate table(s), and create a pull request. We will review your pull request (and your files on zenodo) as soon as possible, and ask question - if needed.
Thank you for your contribution!