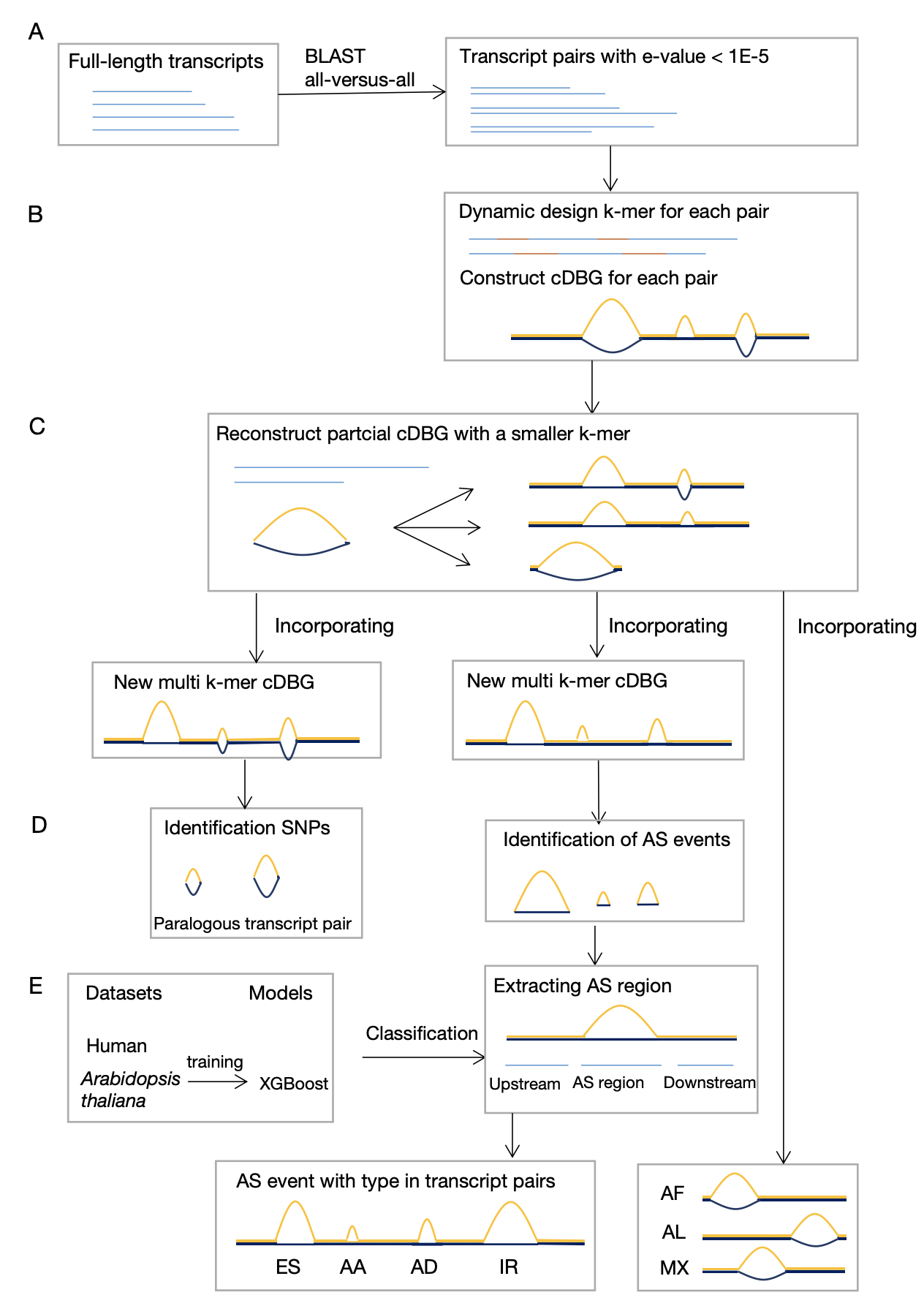
Alternative splicing (AS) is an essential post-transcriptional mechanism that regulates many biological processes. However, identifying comprehensive types of AS events without guidance from a reference genome is still a challenge. Here, we proposed a novel method, mkcDBGAS, to identify all the seven types of AS events without a reference genome using transcriptome alone. MkcDBGAS, modelled by full-length transcripts of human and Arabidopsis thaliana, consists of two modules. In the first module, mkcDBGAS, for the first time, uses a colored de Bruijn graph with mixed k-mers to identify bubbles generated by AS with precision higher than 98.17%, and detect AS types overlooked by other tools. In the second module, to further classify types of AS, mkcDBGAS added the motifs of exons to construct the feature matrix followed by the XGBoost-based classifier with the accuracy of classification greater than 93.40%, which outperformed other widely used machine learning models and the state-of-the-art methods. Highly scalable, mkcDBGAS performed well when applied to Iso-Seq data of Amborella and transcriptome of mouse. MkcDBGAS is the first accurate and scalable method for detecting all seven types of AS events using the transcriptome alone, which will greatly empower the studies of alternative splicing in a wider field.
Dependencies:
Python 3.5–3.8
Biopython
joblib
pandas
numpy
blastn 2.10.1
Installation: After testing all dependencies works well, you can git clone it into your working directory, and all executable file placed in bin/
git clone https://github.com/CMB-BNU/mkcDBGAS.git
cd mkcDBGAS/bin
chmod 777 identifier.sh
echo "export PATH=`pwd`:\$PATH" >>~/.bashrc && source ~/.bashrc
Userage : sh identifier.sh transcript.fasta model thread
transcript.fasta : the full-length transcript in fasta format
model : choosing from [arabidopsis, human], arabidopsis or rice for plant, human for animal
thread : the number of thread to construct the colored de Bruijn graph(cDBG)
All the output files located in the path of input fasta file which prefix is the name of the fasta file