New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Align ticks in at-risk count table #747
Align ticks in at-risk count table #747
Conversation
Update lifelines fork
…labels, explicitly pop kwargs to allow tick labels configuration
|
This is pretty darn exciting! I'll review and test it locally now |
|
Great work, I appreciate the detail PR post as well! In diagnosing this, I've also turned up a few other bugs as well, so thanks for turning my attention to this! |
|
Actually, I did find one bug. If you change the from lifelines.datasets import load_waltons
from lifelines import KaplanMeierFitter, WeibullFitter
from lifelines.plotting import add_at_risk_counts
import matplotlib.pyplot as plt
waltons = load_waltons()
T = waltons['T']
E = waltons['E']
ix = waltons['group'] == 'control'
kmf_control = WeibullFitter()
kmf_control.fit(waltons.loc[ix]['T'], waltons.loc[ix]['E'], label='control')
kmf_exp = WeibullFitter()
kmf_exp.fit(waltons.loc[~ix]['T'], waltons.loc[~ix]['E'], label='exp')
ax = plt.subplot(111)
ax = kmf_control.plot_survival_function(ax=ax)
ax = kmf_exp.plot_survival_function(ax=ax)
add_at_risk_counts(kmf_exp, kmf_control, ax=ax)
plt.show()masterbranch |
|
From what I can tell, it has to do with the Edit: the problem is that we write the labels on the first tick, but this tick location is assigned to 0, and if the lower-bound of xlim is greater than 0, that tick is never shown. A solution is to skip ticks that will never be shown, and then on the first shown tick ( greater than lower-bound), write the labels. Ex code that works: |
|
Hey, thanks for taking a look so quickly! The Weibull fitter example is really useful. I think a good way to cover both cases is to take the Any idea why the |
The xlim automatically set by mpl after we provide mpl the data to plot. In all cases, the x-variable is the |
👍 seems like a good solution |
|
Ok, I changed how the Here is boilerplate code for the plots below. DefaultSetting
|
looks great! Thanks for this attention to detail! |
|
Great! Thanks for your help and great work! |


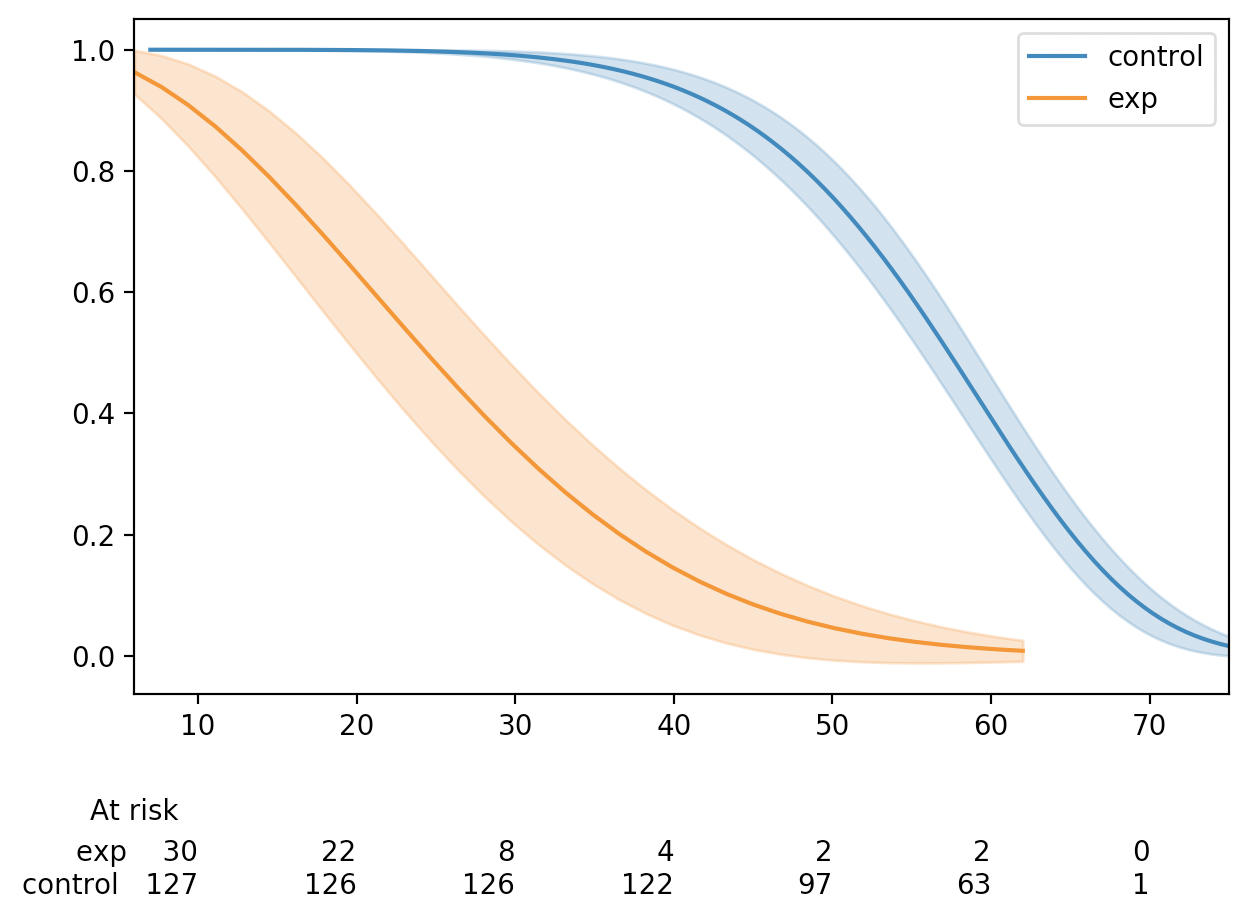
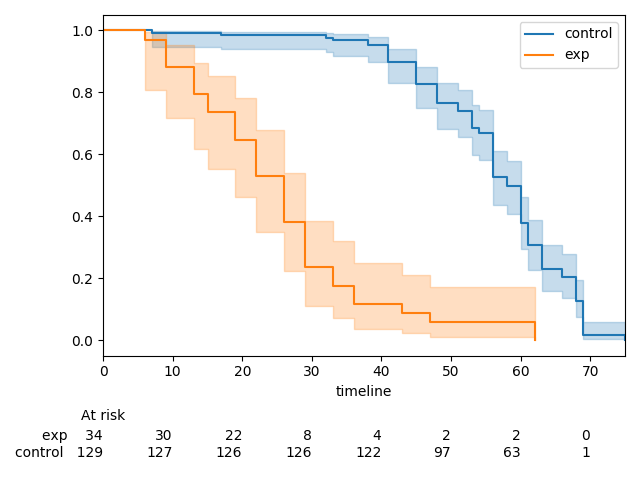
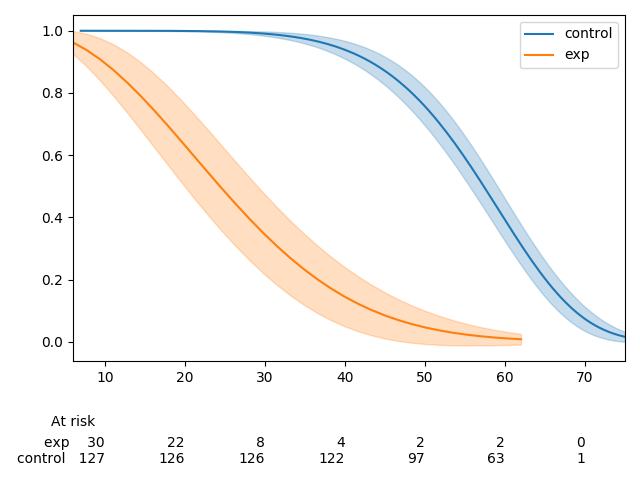

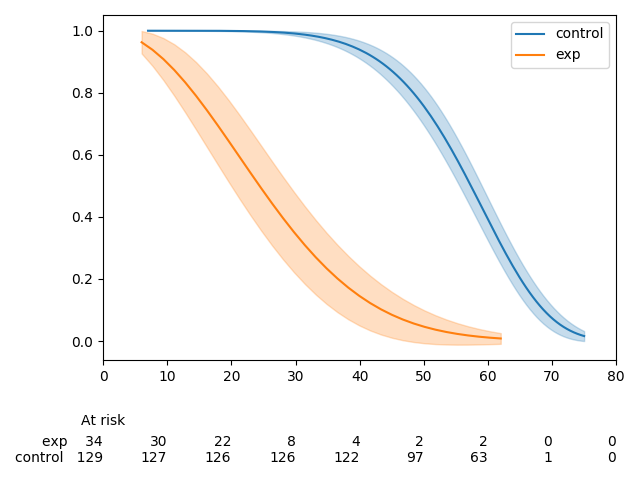
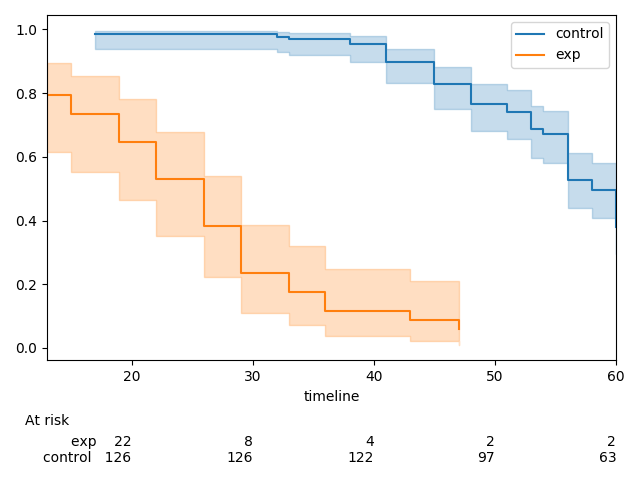
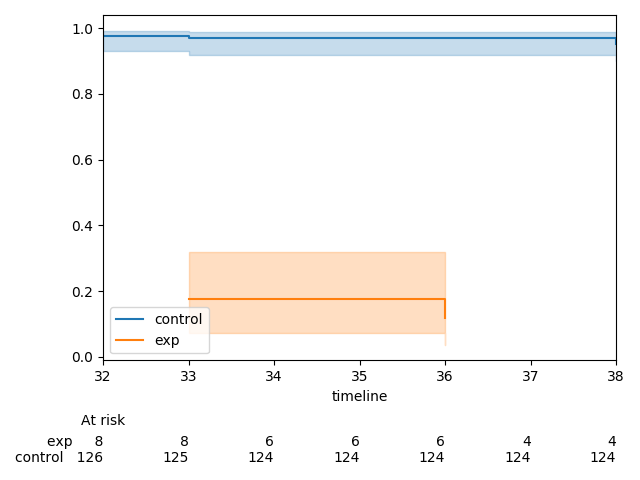
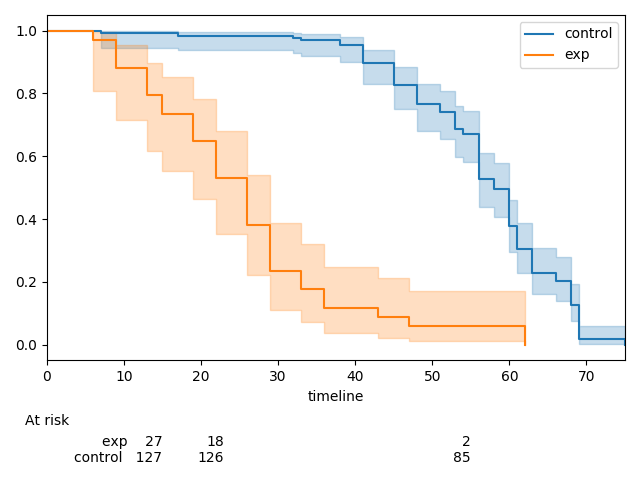


Currently,
add_at_risk_countsgenerates a set of xticks that get added to a separate axes object below the original axes. In some cases the ticks can be mis-aligned due to differences in how thexlimandxticksobjects from the initial axes object being passed in are used to generate the new axes object.The documentation has a good example of this, which can be reproduced with the code below.
There is one extra tick in the at-risk counts than in the plot. This can be fixed by re-ordering how the
xlimandxticksobjects are created.This MR does three things:
kwargsto be passed in toadd_at_risk_countsthat can be passed into theset_xticklabelsmethod (e.g.fontsize)Before
After