A Python-based software for the treatment of X-ray scattering data of amorphous and liquid systems
Boccato S., Garino Y., Morard G., Zhao B., Xu F., Sanloup C., King A., Guignot N., Clark A., Garbarino G., Morand M., Antonangeli D., Accepted in High Pressure Research, Amorpheus: A Python-based software for the treatment of X-ray scattering data of amorphous and liquid systems
https://doi.org/10.1080/08957959.2022.2032032
- Silvia Boccato: silvia.boccato@upmc.fr
- Yiuri Garino: yiuri.garino@cnrs.fr
-
Download and install Anaconda https://docs.anaconda.com/anaconda/install/
-
from anaconda prompt (windows) or terminal (Ubuntu & MAC) add conda-forge channel
conda config --add channels conda-forge
conda config --set channel_priority strict- create virtual environment with name "Amorpheus" and the necessary libraries
conda create -n Amorpheus spyder spyder-kernels xraylib=4.0.0 lmfit matplotlib periodictable ezodf configparserNote: This procedure will install the last Spyder version. For a stable version please use the command indicated in Known issues
- activate the virtual environment with
conda activate Amorpheus - launch spyder with
spyder- open the file Amorpheus.py and run the program.
Test files are provided: Cerium.dat and background.xy. The associated input file is Init.txt. Batch function can be tested with the input file Batch.ods
or
open the file RPS.py and run the program. A test file will automatically be created and visualized.
- close the virtual environment with
conda deactivateIt is possible that the last available version of Spyder in Anaconda is not compatible with a smooth functioning of Amorpheus. To install a stable version of the virtual environment, delete the old environment with
conda env remove -n Amorpheus move to the Amorpheus folder with the file Amorpheus_requirements.txt and substitute step 3. of the installation with the following command
conda create -n Amorpheus --file Amorpheus_requirements.txt- from anaconda prompt (windows) or terminal (Ubuntu & MAC) activate the virtual environment with
conda activate Amorpheus - launch spyder with
spyder- make sure that the folder where you’re performing the analysis contains the following files
- Amorpheus.py : the main software
- equations.py : file of the equations, it is read by the main software
- Init.txt : file with the initial parameters. A detailed description of all the parameters can be found in Parameters
- Batch.ods : file with the initial parameters when using the option Batch [b] in the main menu
- filename.dat : data file, it has to appear with the same name and same extension as in the Init.txt or Batch.ods
- background.xy : file for the background, when applicable. It has to appear with the same name and same extension as in the Init.txt or Batch.ods
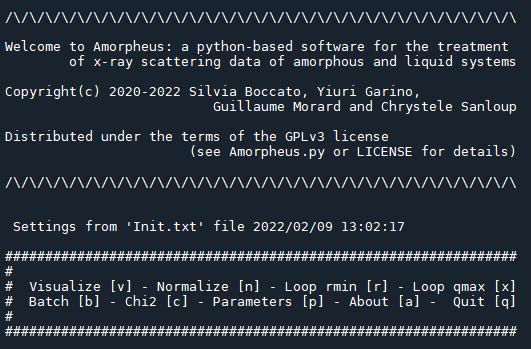
- open the file Amorpheus.py in spyder and run the program. The command-line-based user interface allowing to navigate in the menu will show as:
An example of input file Init.txt is provided for the case of the Ce-based glass shown in Data treatment. The values of all input parameters can be changed also in the section Parameters [p] of the main menu.
-
[1]-[2] Filename - Filename background (str): These parameters are the names of measured
$I^{meas}$ ,$I^{bkg}$ respectively. The$I^{bkg}$ is pertinent only if the File Format supports a background. Columns have to be separated by space or tab (files with comma separated columns will not work). -
[3] File Format (int): Allows to chose four possible file formats:
- File Format = 1 : data are in
$2\theta$ and there is no background - File Format = 2 : data are in
$2\theta$ with a background in the same units - File Format = 3 : data are in
$Q$ in$Å^{-1}$ with no background - File Format = 4 : data are in
$Q$ in$Å^{-1}$ with a background in the same units
- File Format = 1 : data are in
-
[4] incoherent (int): The parameter of incoherence is set to 1 when the background was measured on the empty celland to 0 when the background was measured on the sample. In this case the sum of the incoherent scattering signals from the sample is already subtracted while subtracting the background, and in the normalization the term
$\sum_p I_p^{incoh}(Q)$ is set to 0. -
[5] lambda (float): It is the wavelength
$\lambda$ of X-rays expressed in nm. The software reads this parameter only in case of File Format = 1 and 2, when the conversion from angle$2\theta$ to scattering vector$Q$ is performed as$q = \frac{4 \pi}{\lambda} \sin(\theta)$ . In the case of Cerium parameter lambda is set to 1 as the File Format is 4 and this value is not used. In the case of liquid iron presented in Documentation this parameter was set to lambda = 0.03738. -
[6]-[7] qmin - qmax (float): Parameters qmin and qmax are the limits for the
$Q$ range under analysis, they always have to be expressed in$Q$ even with File Format = 1 and 2. The value of$Q_{min}$ has to be chosen reasonably lower than the position of the diffuse scattering peak, its precise value has little effect in locating the peaks of the radial distribution function. At the first iteration$S(Q)$ is defined between$Q_{min}$ and$Q_{max}$ , but for the aforementioned reason it is typically extrapolated at low$Q$ as$S(Q)=S(Q_{min}), \forall Q : 0 < Q < Q_{min}$ . A thorough choice of$Q_{max}$ is instead significant in the minimization of the figure of merit. The Loop over qmax [x] section of the software helps in the choice of$Q_{max}$ . -
[8] rmin (float): The choice of the cutoff radius
$r_{min}$ is also crucial for the optimization of the figure of merit. This value can be chosen with the help of the Loop over rmin [r] section of the software. This value can take on positive values lower than 0.4 nm. -
[9] bkfactor (float): The bkfactor parameter is the scale factor
$b$ . Its value, at the beginning set to 1, is optimized in the minimization of the figure of merit. In the optimization this value can take on values between 0.7 and 1.2. -
[10] density (float): The value of the atomic density
$\rho_0$ is also optimized during the minimization procedure. It is expressed in$at/nm^3$ and during the optimization it can take on values between -50 and +50$at/nm^3$ with respect to the initial set value. -
[11] Normalization Type (int): If set to 1, leads to the use of the modification function. When Normalization Type = 0 the standard normalization is performed.
-
[12] SofQ (int): If set to 1, at the first iteration
$S(Q) = S(Q_{min})$ for$Q < Q_{min}$ . If set to 0, at the first iteration$S(Q) = 0$ for$Q < Q_{min}$ . -
[13] QStep (int): This parameter currently has to be set to 0 (it will be useful in future developments of the software).
-
[14]-[16] Save filenames (str): Parameters for saving figures and files associated to Normalize [n], Loop over rmin [r] and Loop over qmax [x] respectively.
-
[17]-[18] Elements - Content (list-array): It is possible to consider systems with up to 6 elements and it is sufficient to type the element symbol in parameter [17] with the corresponding content in at% in the following line [18]. A check that the sum of the element's content is 1 (100%) is performed in the software.
More details are available in Documentation
- from anaconda prompt (windows) or terminal (Ubuntu & MAC) activate the virtual environment with
conda activate Amorpheus - move to the folder where RPS.py is saved and launch it with
python RPS.pyNote that for RPS it is not necessary that the file to analyze and the Python code are in the same folder.
A screenshot of the program is shown here below:
Amorpheus: a Python-based software for the treatment of X-ray scattering data of amorphous and liquid systems
Copyright (c) 2020-2022 Silvia Boccato, Yiuri Garino, Guillaume Morard and Chrystele Sanloup
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/.
Remove Peaks and Smooth (RPS) - Support tool for the software Amorpheus to pre-treat the data
Copyright (c) 2020-2022 Silvia Boccato, Yiuri Garino, Guillaume Morard and Chrystele Sanloup
RPS is part of Amorpheus. Amorpheus is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version. Amorpheus is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with Amorpheus. If not, see https://www.gnu.org/licenses/.
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (Grant agreement No. 724690, P.I. Daniele Antonangeli).
The authors would like to thank Carlo Boccato for suggesting the name of this software and Micaela Pinola for help with the graphical layout of RPS tool.