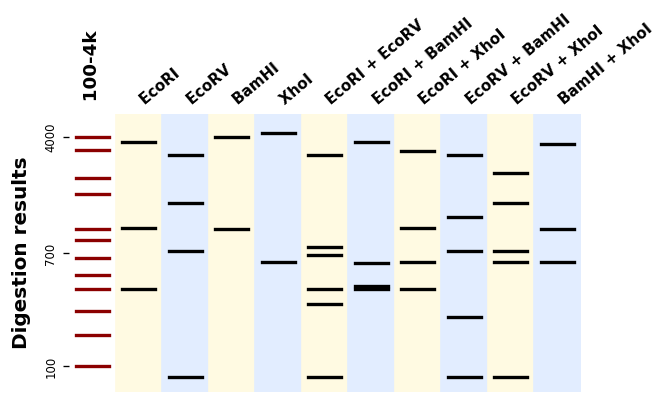
Bandwagon (full documentation here) is a Python library to predict and plot migration patterns from DNA digestions. It supports hundreds of different enzymes (thanks to BioPython), single- and multiple-enzymes digestions, and custom ladders.
It uses Matplotlib to produce plots like this one:
PIP installation:
pip install bandwagonWeb documentation:
https://edinburgh-genome-foundry.github.io/bandwagon/
Github Page
https://github.com/Edinburgh-Genome-Foundry/bandwagon
Live demo
https://cuba.genomefoundry.org/predict-digestions
License: MIT, Copyright Edinburgh Genome Foundry

Bandwagon is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.