ALISTER is a web-app containing scientific information on pre-analytical blood sample stability in metabolomics and lipidomics. With a user-friendly interface and easy navigation, the app provides you with accurate and current information, essential for any researcher or scientist working in these fields. Current app contents are limited to the influences of time delay and temperature during processing of (EDTA) plasma and serum samples, which have been identified to be among the major pre-analytical pitfalls. Within four different menus, the app aims to answer pre-analytical questions regarding two general scenarios. In prospective scenarios the app aims to advise on sampling protocols, that ensure stability of analytes of interest. In retrospective assessment of samples, that ALISTER assesses analytes stability based on pre-analytical sample information provided by the user. Detailed information on the scientific motivation for ALISTER can be found in its publication1. The following document gives an overview of the app functionalities.
ALISTER can provide data for plasma or serum samples. In the first step you need to select which matrix you are working with.
As described above ALISTER is able to answer questions regarding prospective as well as retrospective pre-analytical scenarios. Retrospective assessment of samples in respect to their pre-analytical stability can be made using:
- Sample search
- Analyte search
- Data filtering mode
Prospective planning of sampling can be performed using:
- Protocol search
- Analyte search
Research question: "I have a set of lipids and/or polar metabolites that have been measured from blood samples (e.g. biobank samples). I want to gain information on whether results should be treated with caution due to the mode of sample collection."
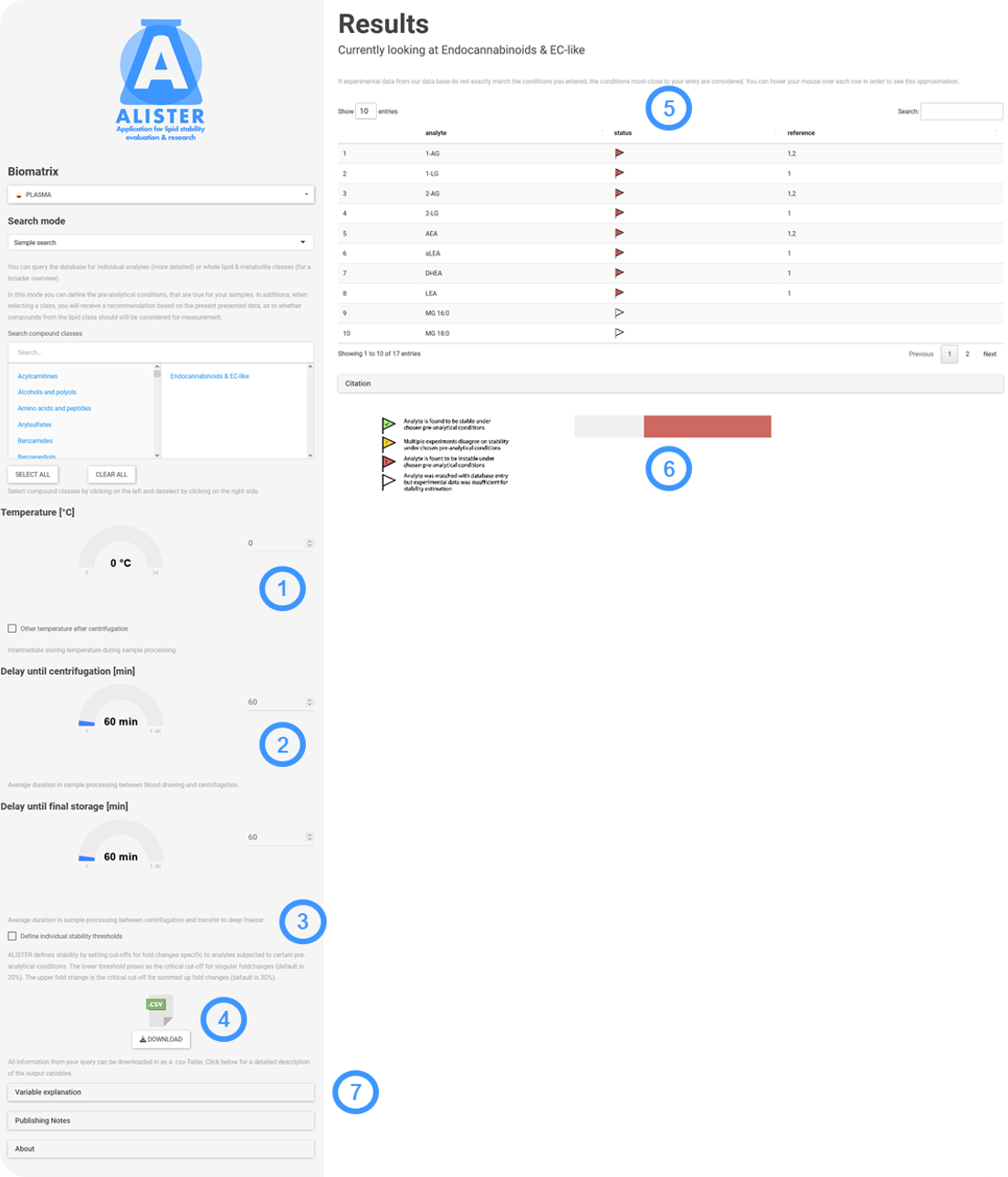
Sample search is used in the following way:
- Select the compound classes of interest from the list. Analytes are categorized by their main lipid class (when lipids) or their main class according to the RefMet database (for polar metabolites). You can choose to 'Select all' groups at once.
- Choose the temperature at which your samples were processed under. If the temperature varied before and after centrifugation, check 'Other temperature after centrifugation' and add the additional information.
- Experiments have shown, that for specific analytes or analyte classes it does matter, whether a time delay occurs before or after centrifugation. Enter the respective processing delays.
- By default the critical threshold for assessing instability is set to 20% fold change due to specific pre-analytical conditions. When conditions are combined (e.g. added change due to delay before and after centrifugation) the threshold is expanded to 30%. You can choose to readjust this upper and lower limit by checking 'Define stability thresholds'.
- This is the result of your query. In some cases, the database underlying ALISTER will not exactly match your conditions. In these cases, generalization needs to happen. Hover your mouse over each analyte in order to see how your input was interpreted. Click on 'Citation' in order to see relevant literature on the stability of the displayed analytes.
- A pie chart shows the overall influence of your input conditions on the set of selected analytes.
- You can download a .csv-file containing your query, as well as the output and all relevant references. The tab 'Variable explanation' will answer all your questions regarding interpretation of the download.
Research question: "I want to take blood samples in order to analyze a certain set of lipids and/or polar metabolites. What is the optimal way of sampling in order to assure stability for most analytes of interest? What is the optimal way in order to ensure stability for all of them?"
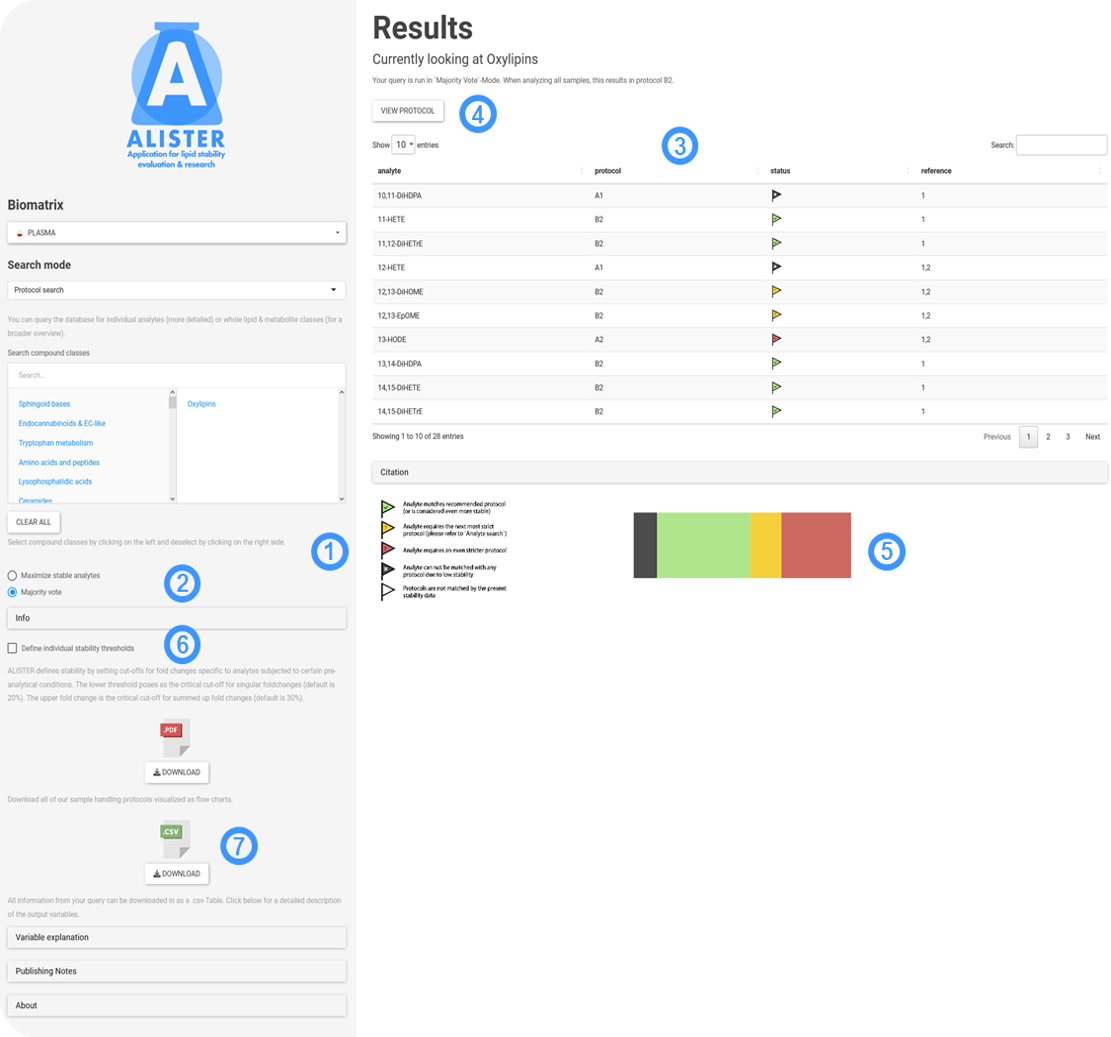
Protocol search is currently only available for plasma samples. It is used in the following way:
- As for the 'Sample search', you can choose the specific analyte classes of interest.
- If you want to receive the sampling protocol that is recommended in most cases for the selected analytes, choose 'Majority Vote' (default). In other cases, you might want to choose the strictest protocol in order to ensure stability for the maximum number of selected analytes. For this, choose 'Maximize stable analytes'.
- The output is straight forward. Based on its inherent pre-analytical stability, each analyte receives a recommended sampling protocol. The overall protocol is recommended above the table. An icon next to the analyte indicates, whether the respective analyte is expected to be stable under the recommended protocol (this is mostly relevant when working in 'Majority Vote').
- You can investigate the recommended sampling protocol in the form of a simple flow chart.
- A pie chart summarizes the expected stability of all selected analytes under the chosen conditions.
- Like previously, for even more refined research questions you can adjust the stability thresholds to be more or less strict.
- In addition to downloading a .csv-file containing your query, as well as the output, you are able to download a PDF file, that depicts all possible sampling protocols in straight forward flowcharts. The tab 'Variable explanation' will answer all your questions regarding interpretation of the download.
Research question: "I am interested in the stability of a very specific analyte. I need detailed information on how to take sample properly, as well as on how stable the analyte is under certain pre-analytical conditions."
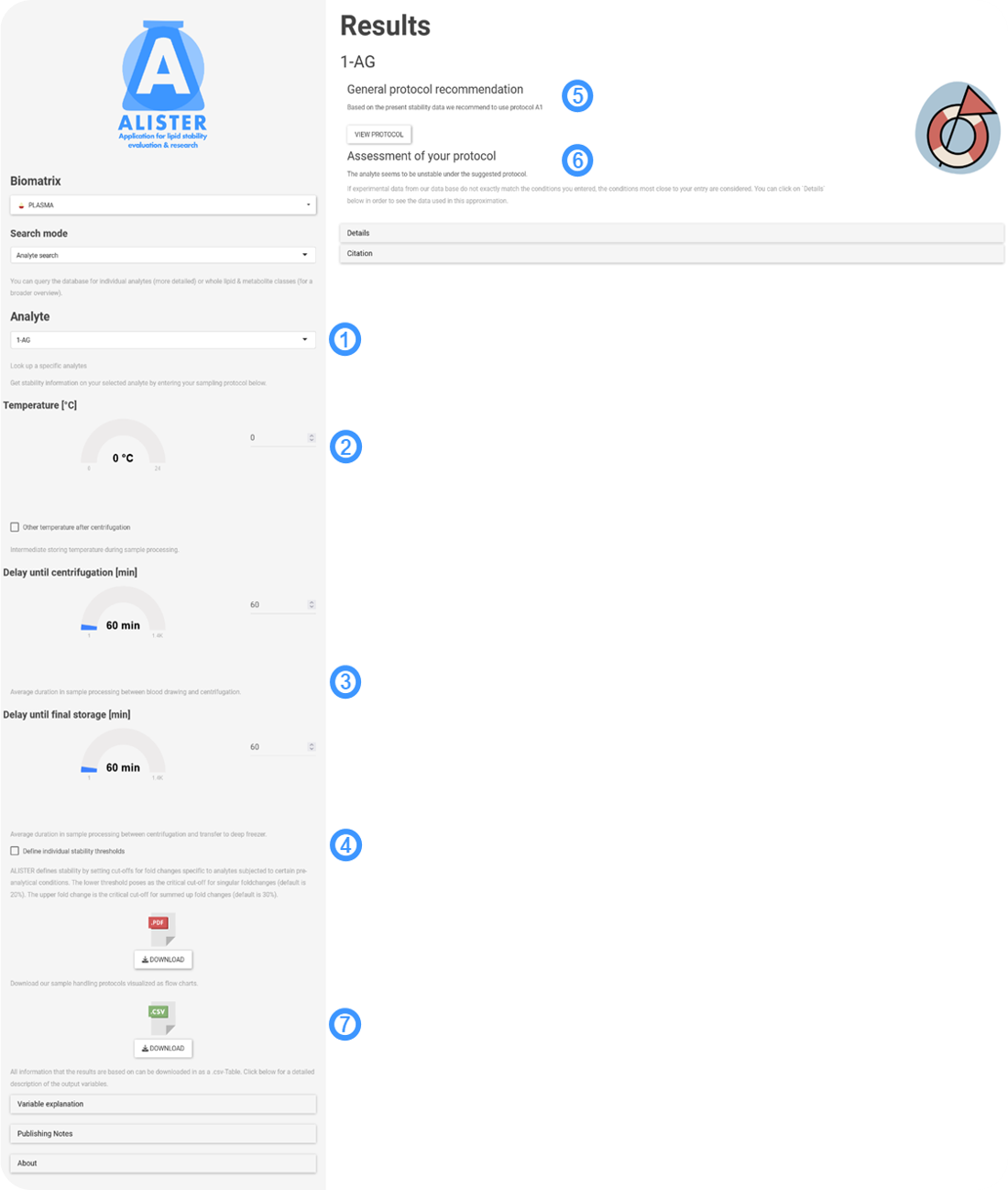
Analyte search is used in the following way:
- As 'Analyte search' provides a more detailed investigation of analytes, you can select them individually in the drop-down menu. It is possible to type in the name of your analyte of interest, so you don't have to scroll through all of them.
- As in 'Sample search' you can now add your pre-analytical conditions, starting with the temperature during processing. Click 'Other temperature after centrifugation' if the processing temperature varied before and after centrifugation.
- Processing delay before and after centrifugation might affect the analyte in different ways. Input the processing time of your samples here.
- As previously: If you want to define more or less strict stability thresholds, you can do that here.
- 'Analyte search' combines functions previously introduced in 'Sample search' and 'Protocol search'. Here, the protocol recommendation, as well as the visual output introduced in 'Protocol search' are displayed.
- Additionally, 'Analyte search' assesses your own pre-analytical input conditions. When clicking on 'Details' you can see a summary of the literature information that ALISTER bases its decision on, a generalized form of your input and whether the set individual threshold of fold changes due to pre-analytical stability was exceeded and in which direction.
- All data you entered in your query together with the respective output can be downloaded as a .csv file. The tab 'Variable explanation' will answer all your questions regarding interpretation of the download.
Research question: "I have a set of measurements of various specific analytes. I do know the conditions under which the samples were taken. I want an indication for which analytes the measurement might be unsuitable due to pre-analytical conditions. I also want to filter them out of my dataset pre-emptively."
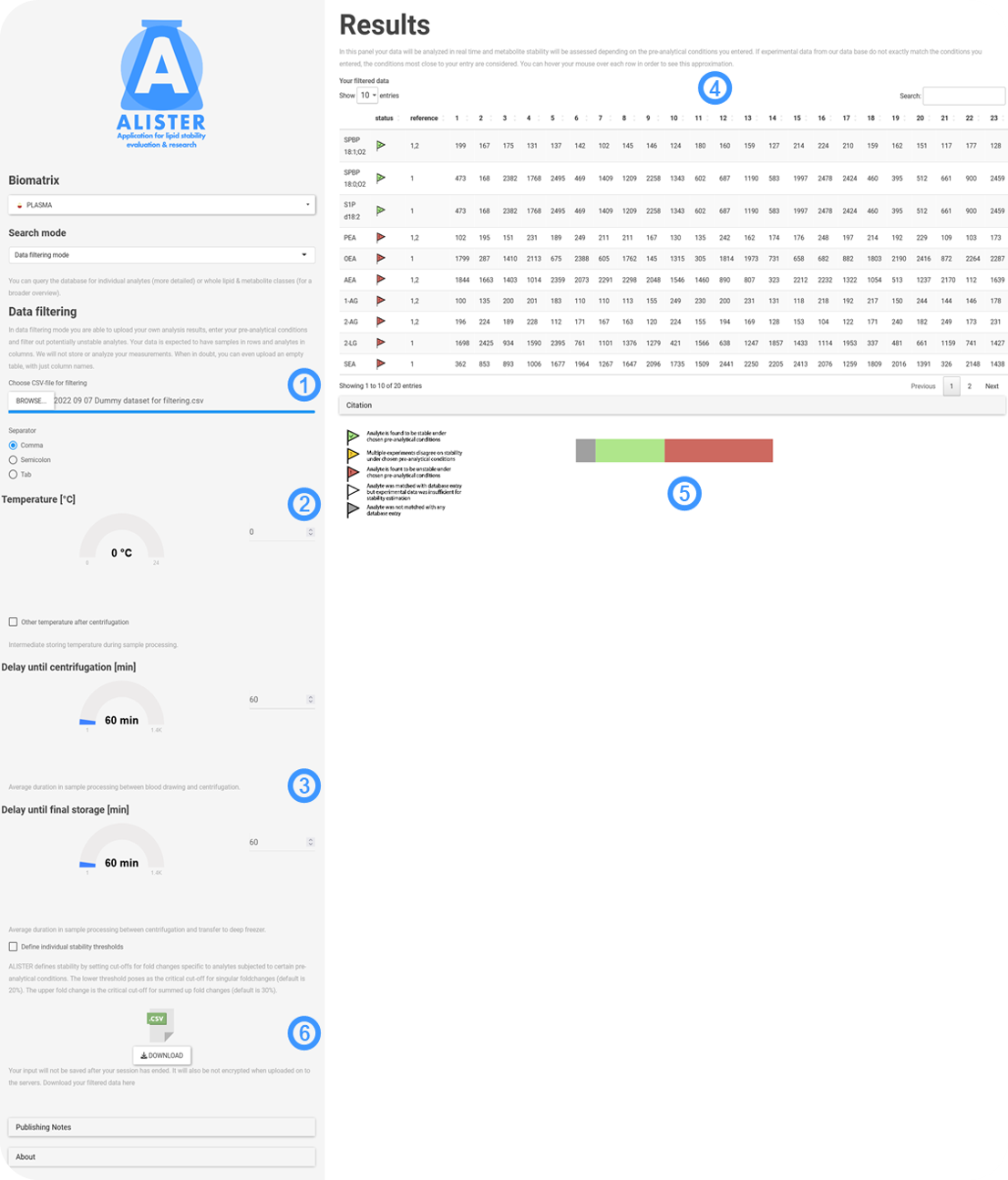
The data filtering mode is used in the following way:
- First up you need to specify a .csv file for upload into the app environment. Despite being displayed in a transposed way, the app requires that the file contains the input data in tabular form, with sample names in the rows and analyte names in the columns. Your data will not be altered or stored. It is even possible to upload an empty table, that contains only the analyte names relevant to your enquiry. The analyte names will be automatically matched against the ones in ALISTER´s underlying database. Lipids are named after the lipid shorthand nomenclature2, while polar metabolites are named after their RefMet nomenclature3.
- As in 'Sample search' you can specify the temperature during processing. Click 'Other temperature after centrifugation' if you want to put in different temperatures for pre- and post-centrifugation processing.
- Input the processing delays before and after centrifugation. In most pre-analytical experiments their influence is tested separately, but here both will be taken into consideration. In this way, pre-analytical influences will be assessed separately and then amalgamated. As in each of the search modes discussed, you can adjust the thresholds for pre-analytical stability individually.
- This is your data. Each analyte is accompanied by a visual indicator giving you a hint on whether the analyte can be expected to be stable under your input conditions based on experimental data from literature. Hover your mouse over each analyte in order to see how your input conditions were approximated by available experimental data. Click 'Citation' if you want to see the sources relevant to assessing the stability of your input.
- A pie chart indicates the percentages of each stability category among your analytes under the given conditions.
- The .csv download is a bit different from previous search modes. Despite still giving you the possibility to save your input conditions, you are presented with an annotated version of your dataset. All analytes, that were evaluated as 'unstable' based on experimental stability data from literature and your input conditions are flagged as such in this variant of your dataset.
ALISTER aims to aggregate analyte stability data and facilitate low-threshold access for the assessment of data and blood sample quality. Such a data-driven approach of course necessitates large amounts of experimental data. If you own data that you think could fit ALISTER or know of a study that you wish to be included in ALISTER´s database, please consider contacting us via lisa.hahnefeld@itmp.fraunhofer.de or robert.gurke@itmp.fraunhofer.de.
Footnotes
-
Future reference ↩