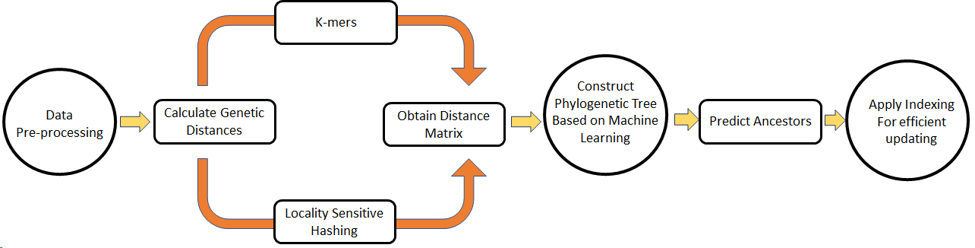
From this research we propose two novel alignment-free, pairwise, distance calculation methods based on k-mers and locality sensitive hashing. In addition to that, we proposed a machine learning-based phylogenetic tree construction mechanism. With the proposed approaches we can gear up the efficiency and accuracy of genetic distance calculation. The tree construction method which is based on a modified version of k medoid is also guaranteed to provide significant performance compared to traditional phylogenetic tree construction methods. As the final part of the research, we implemented a numerical neural network to efficiently update the phylogenetic tree. So in summary with this research, we implemented novel methods of genetic distance calculation, phylogenetic tree construction, and tree updation.
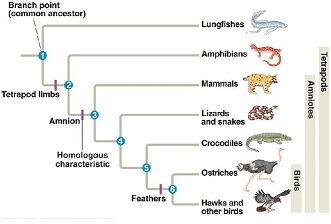
The phylogenetic tree (Evolutionary tree) is a branching diagram that shows the evolutionary relationships among various organisms. It branches out species by considering the similarities of them based on the genetic distance. Phylogenetic tree can be considered as one of the fundamental components of most of the bioinformatic research.
If you are using this system please cite original authors to credit them.
- [1]G. Gamage, N. Gimhana, A. Wickramarachchi, V. Mallawaarachchi, and I. Perera, “Alignment-free Whole Genome Comparison Using k-mer Forests,” in 2019 19th International Conference on Advances in ICT for Emerging Regions (ICTer), 2019, doi: 10.1109/icter48817.2019.9023714.
- [2]T. Pathirana, S. Bandara, G. Gamage, N. Gimhana, A. Wickramarachchi, and I. Perera, “Genetic Distance Calculation based on Locality Sensitive Hashing.” Cold Spring Harbor Laboratory, 07-Apr-2020, doi: 10.1101/2020.04.06.027250.
- Papers accepted not published yet - PHYLOGENETIC TREE CONSTRUCTION USING K-MER FOREST-BASED DISTANCE CALCULATION G. GAMAGE, N GIMHANA , A. WICKRAMARACHCHI , V. MALLAWAARACHCHI , I. PERERA, S. BANDARA, T. PATHIRANA International Journal of Online and Biomedical Engineering (iJOE),Vol 16, No 02 (2020)