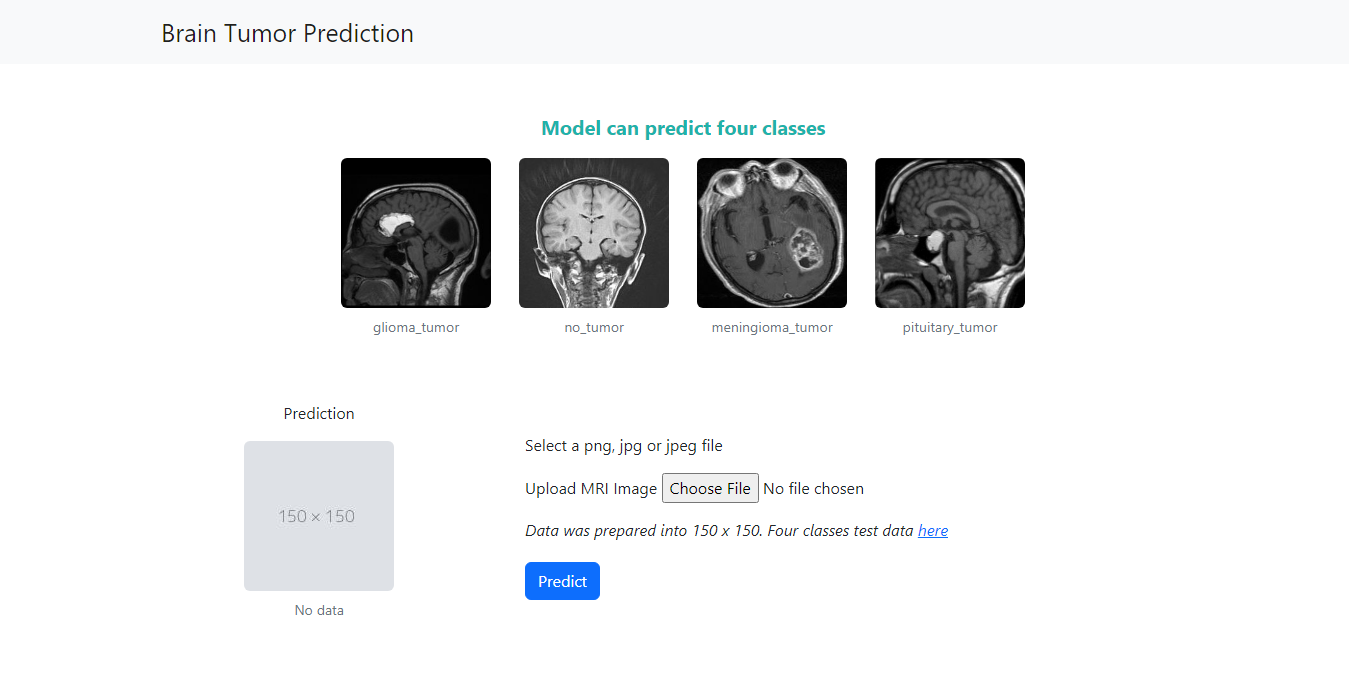
A deep learning model for predicting brain tumor from MRI images using TensorFlow Convolutional Neural Network (CNN). Transfer learning is used to train the model. The model has four classes: meningioma, glioma, pituitary tumor, and no tumor with 98% prediction accuracy.
The model is deployed on here: https://btpred.soleyman.xyz
- TensorFlow - The machine learning framework used
- Keras - The high-level neural networks API used
- EfficientNet - For transfer learning
- Flask - The web framework used
- OpenCV - For image processing
- Matplotlib, Seaborn - For data visualization and analysis
- Clone the project
git clone https://github.com/IMSoley/PredBrainTumor - Install dependencies
pip install -r requirements.txt - Create upload dir
mkdir static/uploads - Run the app
python app.py - Visit
http://127.0.0.1:5000
The model folder contains the trained model. Model testing data is in the testing folder. These data can be used to test the model locally or on the deployed website.
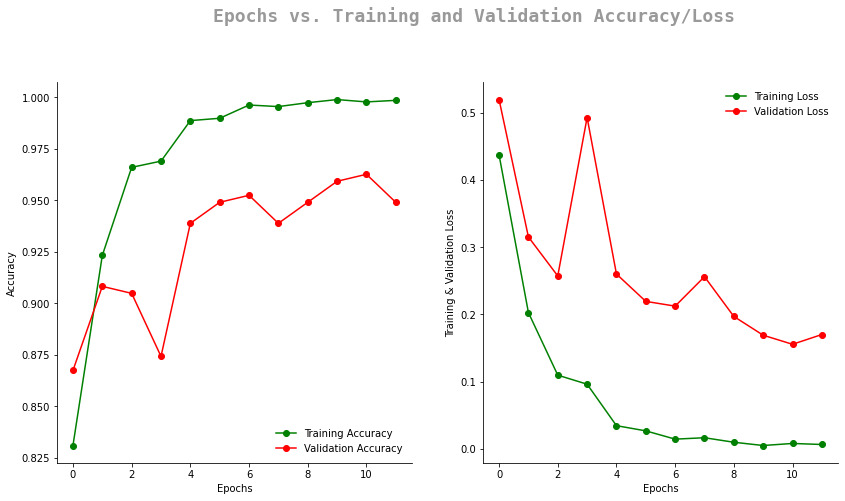
The model is trained on the Brain Tumor Classification (MRI) dataset from Kaggle. The model is trained on 2,937 images and tested on 327 images. The model has 98% accuracy on the test set.
# EfficientNetB0 a convolutional neural network that is trained on more than a million images from the ImageNet database
effnet = EfficientNetB0(weights='imagenet', # using the weights from the ImageNet database
include_top=False,
input_shape=(image_size, image_size, 3))
# effnet output is the input to the model
model = effnet.output
model = tf.keras.layers.GlobalAveragePooling2D()(model)
model = tf.keras.layers.Dropout(rate=0.5)(model)
# this is the output layer with 4 classes
model = tf.keras.layers.Dense(4, activation=tf.keras.activations.softmax)(model)
model = tf.keras.models.Model(inputs=effnet.input, outputs=model) # calling the classification report function
get_classification_report() # calling the confusion matrix function
cm_analysis(y_test_new, prediction, "fig", classes, labels)Note: The model is trained on Kaggle's GPU. It takes approximately 5 minutes to train the model for 12 epochs.
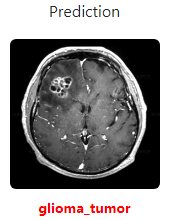
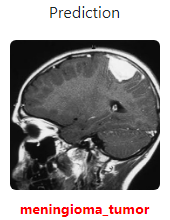
| glioma_tumor(0) | no_tumor(1) | meningioma_tumor(2) | pituitary_tumor(3) |
|---|---|---|---|
 |
 |
 |
 |
MIT License