Drug R+: A comprehensive database for drug repurposing applications
Drug R+ is the first database which provides drug repurposing capabilities based on drug-target interactions, adverse reaction of drugs, and mechanism of action of drugs on targets. Furthermore, expert users can express their complex queries such as nested queries, and then get their favorable results. Also, unprofessional users can state their queries in an easy way. After acquiring the results, they can export them into an excel file.
The first page of Drug R+, which is observer in fgi.1, includes four parts, including Search, CDR, data sets, and database structure.
Fig.1: The first page of Drug R+
In continue, each of the sections is described.
SEARCH
Two search strategies are available in Drug R+. In the first strategy, unprofessional users can select their tables and can add some constraints which limit search space. When a user selects a table and adds a constraint by clicking on the “add constraint button”, the relevant query is built. For acquiring the result, users must click on “show results” button. The first condition is started by “where” while others begin with “and”. If your results are not presented, click on “clear” button and try again. By clicking on “export to excel” button, users can send their obtained results into an excel file. Various parts of the search section are depicted in fig.2. Only, the queries, which are relative to information retrieval, are permitted. In contrast, delete, update, insert, or any other operations which change the database are not allowed.
Fig.2: The search section of Drug R+
CDR
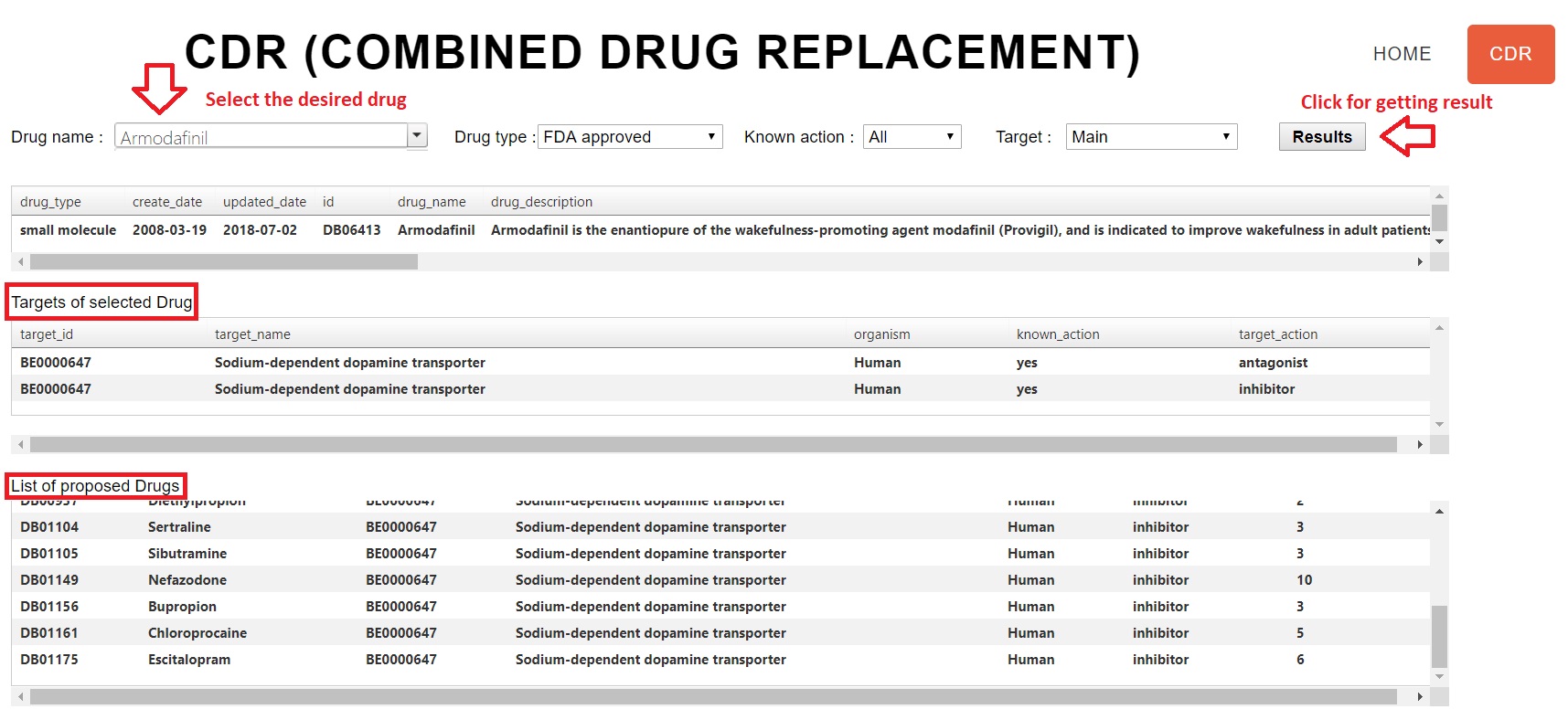
In addition to the search section, CDR section is another main part of Drug R+. This section suggests a list of drugs which have a meaningful relation with a desired drug. In the section, a user must select its desired drug by clicking on the determined field. Then, the user must select the target and click on the “result” button. In the target box, main targets, enzymes, transporters, and carriers can be selected. Also, the user can confine search space using each of the following options:
-
Drug type: users can limit search space for FDA approved or the FDA not approved drugs.
-
Known action: “yes”, “no”, and “unknown” indicate targets which are directly related to their clinical targets, targets which are relative to off-targets, and targets which their mechanism of actions are not reviewed respectively.
After acquiring the list, users must analyze it and obtain their favorable results if they exist. In fig.3, CDR section of Drug R+ is shown. The results of fig.3 are relative to armodafinil which is used for treating excessive daytime sleeping.
Fig.3: CDR of Drug R+
Datasets
Drug R+ includes four datasets which can be used for creating a model for prediction drug-target interactions. In supplementary file, three machine learning approaches have been applied on them, and their results have been reported. The mentioned datasets are:
-
Enzymes: Enzymes are macromolecular catalyzers which accelerate chemical reactions.
-
Ion channel proteins: They are pore-forming proteins which allow ions to pass pore channel.
-
GPCR: G protein coupled receptors are large class of proteins which activate signal transduction pathways.
-
Nuclear receptors: They are responsible for certain type of molecules like steroid and thyroid. In response, they regulate expression of some genes.
All of the mentioned proteins play main roles in a cell and have importance roles in drug design. In fig.4, the dataset section of Drug R+ is presented.
Fig.4: The datasets of Drug R+
DB structure
In order to develop Drug R+, several steps have been followed. In the first step, the flat file of drugs is taken from drug bank database. The flat file is then divided into small files using python programing language. Based on our analysis on the files, we done semantic modeling and then implemented the database. Through third normal form (3NF), the database normalization is done. The script of the database is available in the DB structure section of Drug R+. After that, we used python programing language, and employed parallel processing and map reduce method for transferring data of the flat file into the database. The python codes are also available in this part of web interface of Drug R+. Furthermore, entity relationship diagram (ERD) of the database is accessible. In fig.5, DB structure section is seen.
Users can get a copy of the database by following the below steps:
-
Download the flat file from drug bank. (https://www.drugbank.ca/releases/latest)
-
Divide the flat file into small files using python codes.
-
Create a database using DDL.
-
Run python code for transferring data into their relevant tables and fields.
Fig.5: Structure files of Drug R+
Examples
In this section, several SQL queries and their concepts exist. These examples have different roles and combining them can lead to various results.
-
Getting a total number of drugs
Select count(*) from drugs
-
Getting the drugs which their names include ‘acid’.
Select *from drugs where drug_name like '%acid%'
-
Acquiring a list of drugs which had been created in 2010.
Select *from drugs where DATEPART(year,create_date)=2010
-
The drugs which are FDA approved
Select drug_name from drugs where id in (select id from drug_properties where FDA_label<>'unknown')
-
A list of drugs and a number of their main targets
Select drug_id,drug_name, count(*) from drugs,drug_targets where id=drug_id group by drug_id,drug_name
-
A list of drugs which a number of their targets is greater than 5
Select drug_id,drug_name, count(*) from drugs,drug_targets where id=drug_id group by drug_id,drug_name having count(*)>5
-
A list of drugs which have common enzyme target
Select a.drug_id,b.drug_id,a.enzyme_id,a.enzyme_name from drug_enzymes as a inner join drug_enzymes as b on a.enzyme_id=b.enzyme_id and a.drug_id<>b.drug_id
-
A list of references of a drug with id DB00070 which are relative to its carriers
select *from drug_carrier_articles where drug_id='DB00070'
-
Obtaining a merged list of enzyme and main targets of a drug with id=’DB00005’
(select drug_id as id,target_id as t_id, target_name as target_name from drug_targets where drug_id='DB00005')union
(select drug_id as id,enzyme_id as t_id,enzyme_name as target_name from drug_enzymes where drug_id='DB00005')
-
A list of drugs which are FDA approved, are relative to human organism, have not toxicity, and have common transporter targets with other drugs based on the same mentioned conditions.
select a.drug_id,a.transporter_name,b.drug_id,b.transporter_name from drug_transporters as a inner join drug_transporters as b
on a.drug_id<>b.drug_id and a.transporter_name=b.transporter_name and a.organism=b.organism
and a.drug_id in (select id from drug_properties where FDA_label <>'unknown' and toxicity='null')
and b.drug_id in (select id from drug_properties where FDA_label <>'unknown' and toxicity='null')