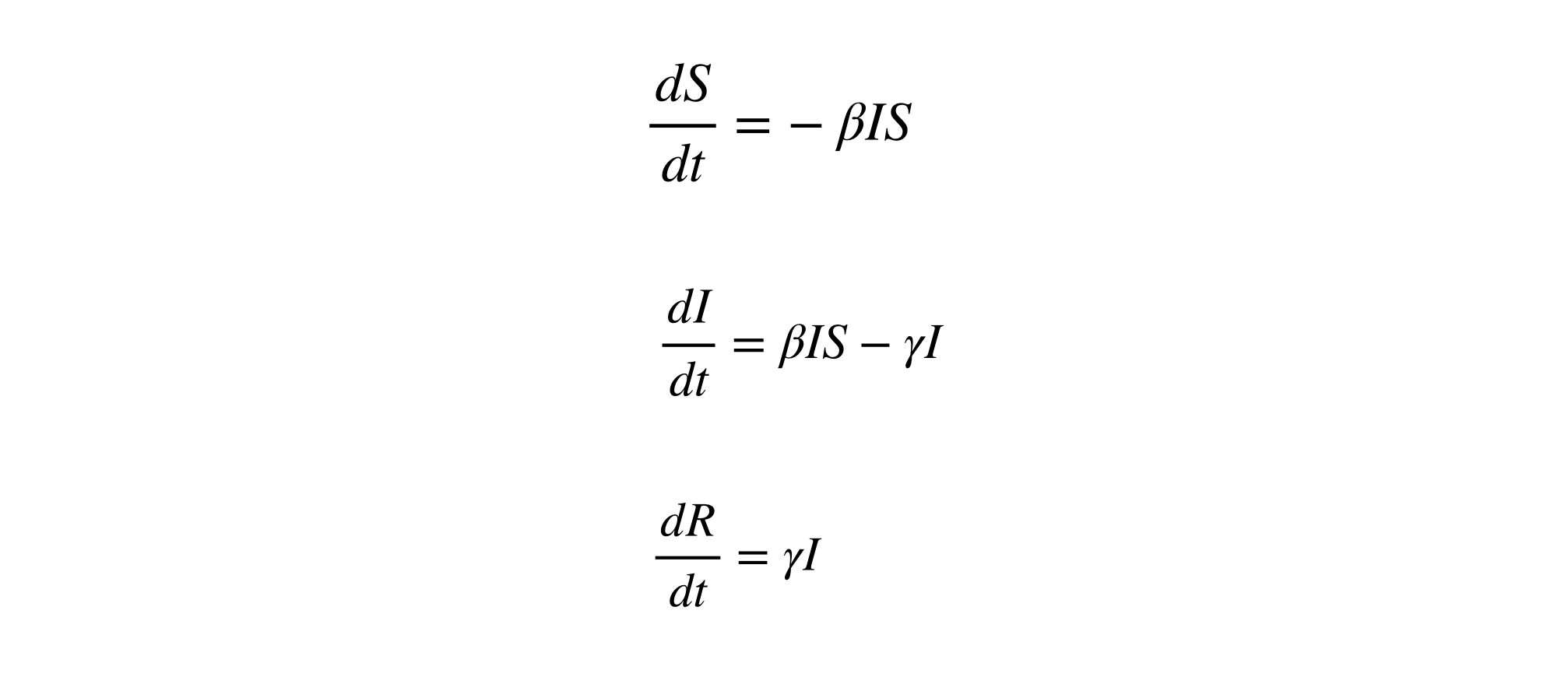COVID19 Modeling aims to create a statistical model simulating the spread of COVID-19 over time. Our model is built based on Susceptible-Infected-Recovered (SIR) models, which is one of the most common techniques in epidemic modeling. This project focuses on developing and examining the effects of different parameters on the basic and extended SIR models in order to understand the complexities of COVID-19 in different scenarios: with and without intervention measures, as well as to underpin such models with real world data to understand their predictive capabilities and limitations.
This research project is being conducted at the Applied Biostatistics Lab of Professor Shonda Kuiper, Grinnell College during Spring 2021.
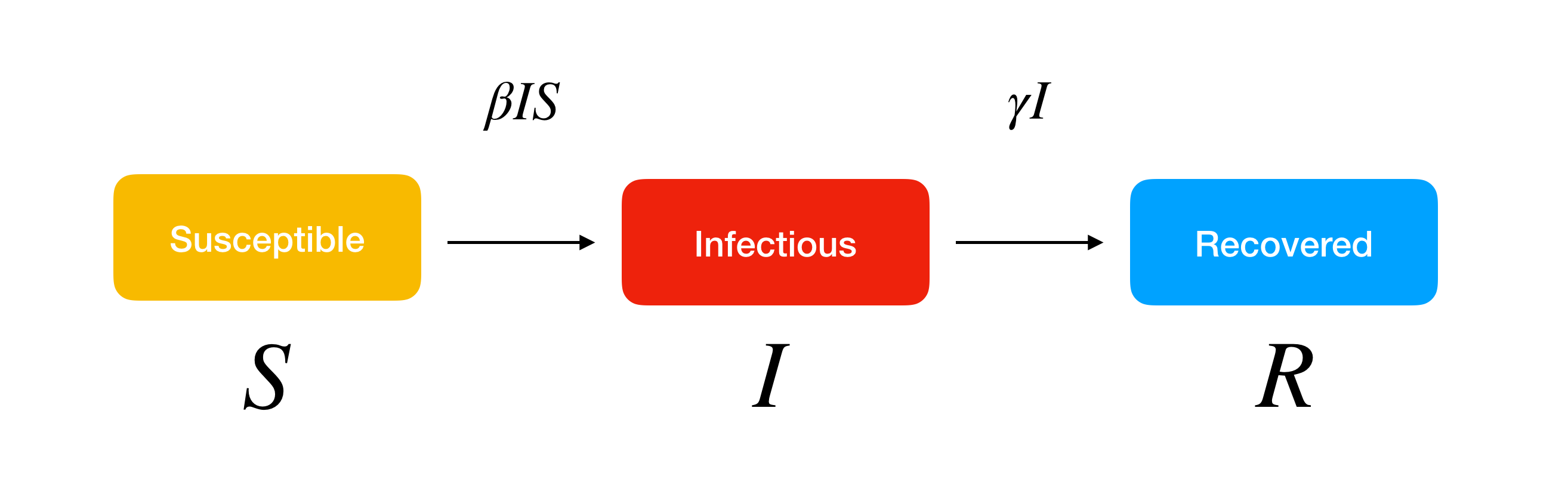
In the basic SIR model, as susceptible members (S) of the population are exposed to a pathogen via an infected individual (I), they are moved to the infectious compartment by an infection rate, and to the recovered/removed compartment (R) after recovery/death by a recovery rate. Many simplifying assumptions are made in developing this model that could change the spread rate of an infectious disease.
The SIR model is based on differential equations, where the fraction of the population in each compartment changes as a function of time. Thus, the functions can be altered depending on the specific disease mechanism – incubation period of the pathogen, the mode of transmission, etc.
The core formulas to predict each compartment S, I, and R over time are as below:
The parameters used in these differential equations, namely the transmission and recovery rates (β & γ) are estimated based upon several factors, including but not limited to the migration rates of the population within and between countries, and the biological mechanism through which the disease affects individuals (Tang et al. 2020). For diseases like COVID-19, where the disease can be both symptomatic and asymptomatic, the models based on SIR can be extended to involve more compartments and levels to capture the complexity of transmission, which is also the ultimate goal of this project.
The source code can be found in this repository. Please note that the project is still in progress of development.
Pull requests are welcome. For major changes, please open an issue first to discuss what you would like to change.
The SIR Model for Spread of Disease - The Differential Equation Model
A mathematical model for simulating the phase-based transmissibility of a novel coronavirus