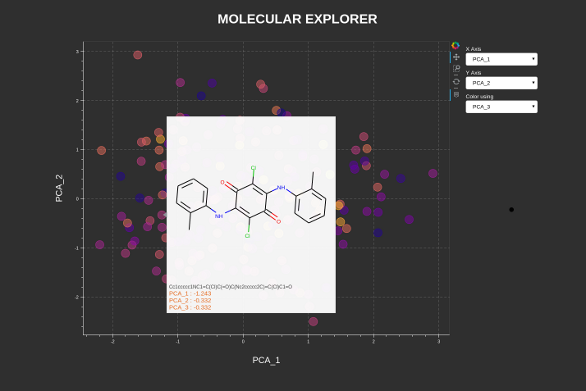
- Choose data column for plot axes and colouring
- Hover over a datapoint to display the structure
- Select datapoints to filter down the data table
- Download selected structures
"bokeh serve . --args [path_to_SMILES_file]"
Need to use --args to provide file path argument through bokeh serve, to the main.py script.
The SMILES file should have a first column containing SMILES, and 2 or more following columns containing scalar data/properties. The first row should be a head with data/property names. E.g:
SMILES PROPERTY_NAME_1 PROPERTY_NAME_2 PROPERTY_NAME_3
CCC(O)C 1.23 3.4 0.45
c1cccc1 0.76 3.6 0.99
- Python 3
- Bokeh == 0.12.6 (the custom coffee script depends on a module structure that was changed in 0.12.6)
- RDKit
- Numpy