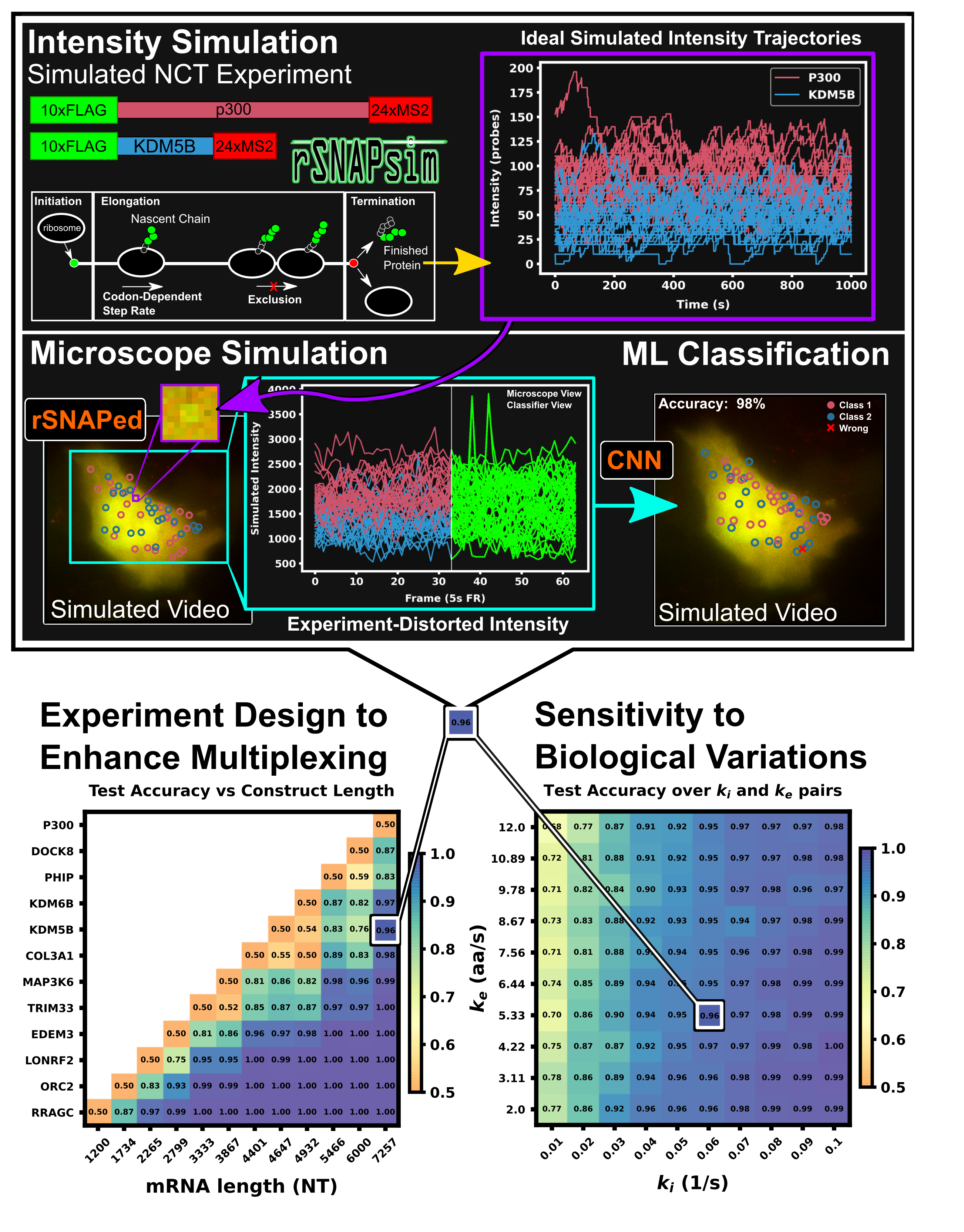
Using mechanistic models and machine learning to design single-color multiplexed Nascent Chain Tracking experiments
William S. Raymond1, Sadaf Ghaffari2, Luis U. Aguilera3, Eric M. Ron1, Tatsuya Morisaki4, Zachary R. Fox1,5, Michael P. May1, and Timothy J. Stasevich4,6, Brian Munsky1,3,*
- School of Biomedical Engineering, Colorado State University, Fort Collins, Colorado, USA
- Department of Computer Science, Colorado State University, Fort Collins, Colorado, USA
- Department of Chemical and Biological Engineering, Colorado State University, Fort Collins, Colorado, USA
- Department of Biochemistry and Molecular Biology, Colorado State University, Fort Collins, Colorado, USA
- Computational Sciences and Engineering Division, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA
- Cell Biology Unit, Institute of Innovative Research, Tokyo Institute of Technology, Nagatsuta-cho 4259, Midori-ku, Yokohama, Japan
For questions about the codes, please contact: wsraymon@rams.colostate.edu, Luis.aguilera@colostate.edu and brian.munsky@colostate.edu
This repository contains the codes necessary to reproduce figures from the above manuscript. All codes are implemented in Python 3.7. Environments used are stored in ./ML_experiments/ML_env.txt and ./datasets/data_making_env.yml. Large data sets and machine learning classifier model files are not stored in this repository but are available upon request.
Using mechanistic models and machine learning to design single-color multiplexed Nascent Chain Tracking experiments, Raymond et al. (Jan 2023) biorxiv preprint
The data sets and code to remake the data are stored in ./datasets .
Data making codes
make_all_data.py- Script that regenerates all simulated data for the machine learning experiments in the paper. THIS WILL GENERATE 120 gb OF DATA! Run at your own risk.run_rsnaped.py- this code runs rSNAPed to generate simulated cells w/ various settings.
Analysis codes
get_diff_pixels.py- calculate diffusion rates of all data sets in the paperget_snrs.py- calculate Signal to Noise Ratios (SNR) of all data sets in the papermatch_particles.py- match particles coming out of tracking for the photobleaching experiment, generate intensity arrays to be used for training in the "realistic tracking condition"get_example_data.py- get some sample intensities / autocorrelations for plotting purposes in the paper.
In the ./ML_experiments folder:
Running machine learning experiments
run_all_ML.py will rerun every ML experiment listed in the paper
Machine learning results
* parsweep_cl_ML: construct length vs construct lengths
* parsweep_img_ML: number of frames vs frame interval
* parsweep_keki_ML: elongation rate vs initation rate for both mRNAs
* parsweep_kes_ML: elongation rate mRNA1 vs elongation rate mRNA2
* parsweep_kis_ML: initation rate mRNA1 vs initation rate mRNA2
* plots: plotted results of the parameter sweep
* metadata.yaml: generation parameters
within each sub folder is
* acc_mat_*_.npy: test accuracy matrix across the parameter sweep
* *_key.csv: test accuracy matrix with row and column labels (pandas)
./ML_F_kis_diff: Frequency only classifier applied to varying imaging conditions
./ML_IF_kis_diff: Intensity + Frequency classifier applied to varying imaging conditions
./ML_I_kis_diff: Intensity only classifier applied to varying imaging conditions
./ML_run_1280_10s_wfreq: Classifier applied to 1280s of NCT for 50 cells video with a 10s frame interval (128 frames, 10s fi) for all comparison analyses
./ML_run_1280_5s_wfreq: Classifier applied to 1280s of NCT video for 50 cells with a 10s frame interval (256 frames, 10s fi) for all comparison analyses
./ML_run_3000_2s_wfreq: Classifier applied to 3000s of NCT video for 50 cells with a 10s frame interval (1500 frames, 2s fi) for all comparison analyses
./ML_run_320_5s_wfreq: Classifier applied to 320s of NCT video for 50 cells with a 10s frame interval (64 frames, 5s fi) for all comparison analyses
./ML_run_tag_3prime: Classifier applied to alternate tag design, 10xFLAG on the 3' end KDM5B vs 10xFLAG p300
./ML_run_tag_base: Classifier applied to original tag design, 10xFLAG on the 5' end KDM5B vs 10xFLAG p300
./ML_run_tag_minus5: Classifier applied to alternate tag design, 5xFLAG on the 5' end KDM5B vs 10xFLAG p300
./ML_run_tag_plus5: Classifier applied to alternate tag design, 15xFLAG on the 5' end KDM5B vs 10xFLAG p300
./ML_run_tag_split: Classifier applied to alternate tag design, 5xFLAG on the 5' end and 5xFLAG on the 3' end KDM5B vs 10xFLAG p300
./ML_PB: Classifiers applied 11 different photobleaching rates of the P300 vs KDM5B default settings
./scripts/ Contains all the scripts used to call run the classification code and return the accuracy matrices over given parameter sets.
Figures and figure generating codes
./figures_data: saved data and codes to recreate each figure
./figures: constructed figures in various formats
Most data files and ML classifiers are not stored on this github (~200 gb), but are available upon request, all data can be regenerated / resimulated from the make_all_data.py script. Any ML experiment can be reran with the run_all_ML.py script.