We propose a novel computational framework EP2vec to assay three-dimensional genomic interactions. We first extract sequence embedding features, defined as fixed-length vector representations learned from variable-length sequences using an unsupervised deep learning method. Then, we train a classifier to predict EPIs using the learned representations in supervised way. Experimental results demonstrate that EP2vec obtains F1 scores ranging from 0.841 to 0.933 on different datasets, which outperforms experimental features-based methods and sequence-based methods. We prove the robustness of sequence embedding features by carrying out sensitivity analysis. Besides, we identify motifs that represent cell line-specific information through analysis of the learned sequence embedding features by adopting attention mechanism. Last, we show that even superior performance with F1 scores 0.889~0.940 can be achieved by combining sequence embedding features and experimental features, which indicates the complementation of these two types of features. In conclusion, EP2vec sheds light on feature extraction for DNA sequences of arbitrary lengths, provides a powerful approach for three-dimensional interactions identification and finds significant motifs through interpreting sequence embedding features. In "EP2vec: a deep learning approach for extracting sequence embedding features to predict enhancer-promoter interactions", we
- Use Paragraph Vector to train enhancer and promoter sequence embedding features seperately in an unsupervised way
- Identify the true enhancer-promoter interactions from other possible interactions in a supervised way
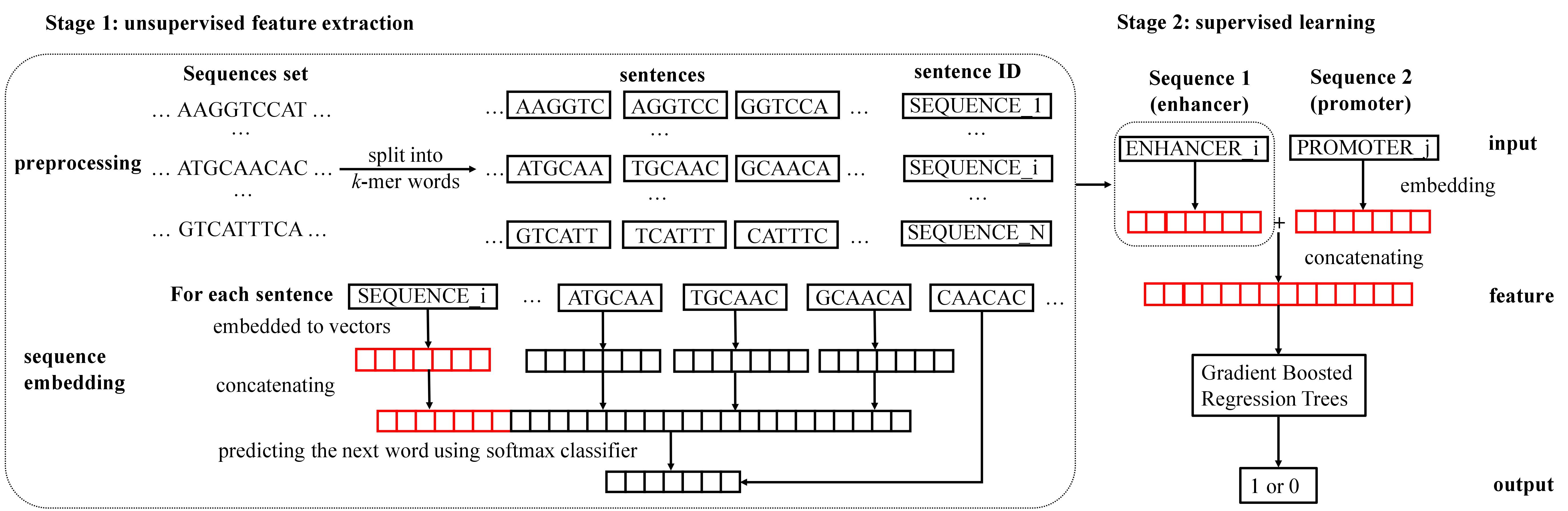
The two-stage workflow of EP2vec. Stage 1 of EP2vec is unsupervised feature extraction which transforms enhancer sequences and promoter sequences in a cell line into sequence embedding features separately. Given a set of all known enhancers or promoters in a cell line, we first split all the sequences into k-mer words with stride s=1 and assign a unique ID to each of them. Regarding the preprocessed sequences as sentences, we embed each sentence to a vector by Paragraph Vector. Concretely, we use vectors of words in a context with the sentence vector to predict the next word in the context using softmax classifier. After training converges, we get embedding vectors for words and all sentences, where the vectors for sentences are exactly the sequence embedding features that we need. Note that in sentence ID, SEQUENCE is a placeholder for ENHANCER or PROMOTER, and is the total number of enhancers or promoters in a cell line. Stage 2 is supervised learning for predicting EPIs. Given a pair of sequences, namely an enhancer sequence and a promoter sequence, we represent the two sequences using the pre-trained vectors and then concatenate them to obtain the feature representation. Lastly, we train a Gradient Boosted Gradient Trees classifier to predict whether this pair is a true EPI.
EP2vec uses the same training data as TargetFinder, where interacting enhancer-promoter pairs are annotated using high-resolution genome-wide Hi-C data (Rao et al., 2014). Labeled training datasets used in the TargetFinder are available in https://github.com/shwhalen/targetfinder.git. We downsample the training sets to ratio 1:1 in all cell types. The K562train.csv, GM12878train.csv, NHEKtrain.csv, IMR90train.csv HUVECtrain.csv and HeLa-S3train.csv are the downsampled training sets. Besides, we need all known enhancers and promoters in a cell line to perform unsupervised feature extraction. We use the enhancers and promoters and TargetFinder in https://github.com/shwhalen/targetfinder.git. The following table show details of each cell line dataset. The enhancers (or promoters) column indicates the number of all known active enhancers (or promoters) for each cell line, which are used for unsupervised feature learning for enhancer (or promoter) sequences.
| Dataset | enhancers | promoters | true EPIs | false EPIs |
|---|---|---|---|---|
| K562 | 82806 | 8196 | 1977 | 1975 |
| IMR90 | 108996 | 5253 | 1254 | 1250 |
| GM12878 | 100036 | 8453 | 2113 | 2110 |
| HUVEC | 65358 | 8180 | 1524 | 1520 |
| HeLa-S3 | 103460 | 7794 | 1740 | 1740 |
| NHEK | 144302 | 5254 | 1291 | 1280 |
ep2vec.py accepts 4 parameters, the length of k-mer, the length of stride, the dimension of embedding vector and the interested cell line. If you want to get the result of 6-mer with stride 1 and embedding dimension 100 in K562, simply run
python ep2vec.py 6 1 100 K562
and you can get the auROC scores, F1 scores and auPRC scores of 10-fold cross-validation. More detailed of the source code can be found in ep2vec.py.
We compared the performance of four methods, namely EP2vec, TargetFinder, gkmSVM, and SPEID, in different datasets, respectively. We rewrite the source code of TargetFinder and SPEID according to their papers in targetfinder.py and speid.py. On the whole, the F1 scores for six cell line datasets of the above four methods range from 0.867 - 0.933, 0.844 - 0.922, 0.731 - 0.822, 0.809 - 0.900, respectively.
EP2vec requires:
- Python
- gensim
- scikit-learn
- bedtools