Simple viewers for ephys signals, events, video and more
Documentation | Release Notes | Issue Tracker
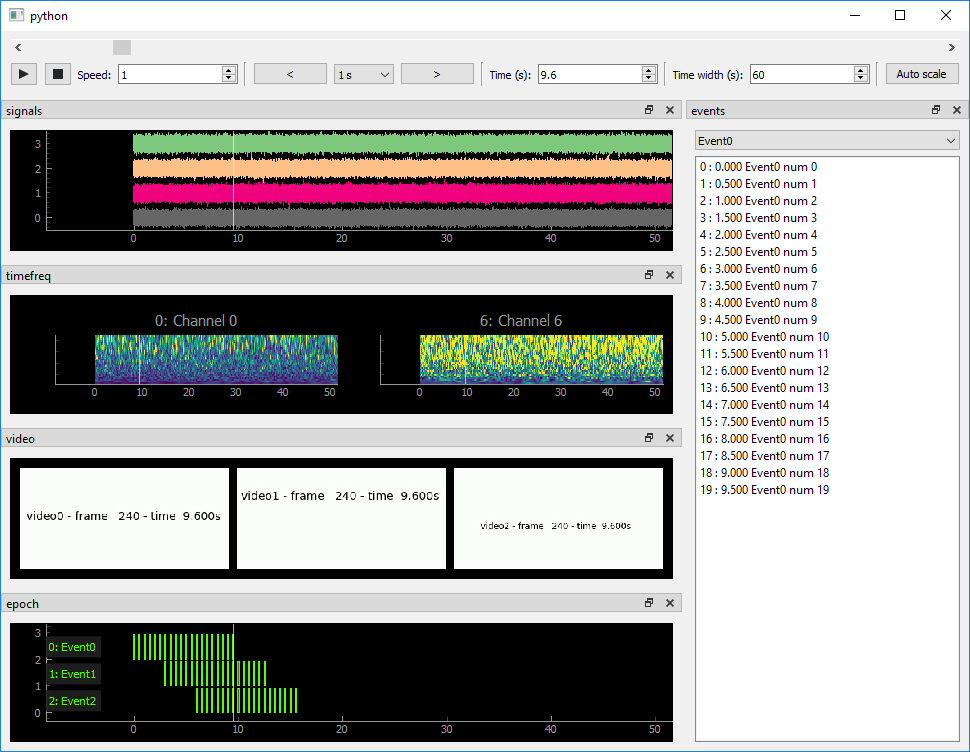
ephyviewer is a Python library based on pyqtgraph for building custom viewers for electrophysiological signals, video, events, epochs, spike trains, data tables, and time-frequency representations of signals. It also provides an epoch encoder for creating annotations.
ephyviewer can be used at two levels: standalone app and library.
For an example of an application that utilizes ephyviewer's capabilities as a library, see the neurotic app and this paper:
Gill, J. P., Garcia, S., Ting, L. H., Wu, M., & Chiel, H. J. (2020). neurotic: Neuroscience Tool for Interactive Characterization. eNeuro, 7(3). https://doi.org/10.1523/ENEURO.0085-20.2020
The standalone app works with file types supported by Neo's RawIO interface (Axograph, Axon, Blackrock, BrainVision, Neuralynx, NeuroExplorer, Plexon, Spike2, Tdt, etc.; see the documentation for neo.rawio for the full list).
Launch it from the console and use the menu to select a data file:
ephyviewerAlternatively, launch it from the console with a filename (and optionally the format):
ephyviewer File_axon_1.abf
ephyviewer File_axon_1.abf -f AxonBuild viewers using code like this:
import ephyviewer
import numpy as np
app = ephyviewer.mkQApp()
#signals
sigs = np.random.rand(100000,16)
sample_rate = 1000.
t_start = 0.
view1 = ephyviewer.TraceViewer.from_numpy(sigs, sample_rate, t_start, 'Signals')
win = ephyviewer.MainViewer(debug=True, show_auto_scale=True)
win.add_view(view1)
win.show()
app.exec()Check the docs for more examples.