Distributed learning for heterogeneous clinical data with application to integrating COVID-19 data across 230 sites
- Description
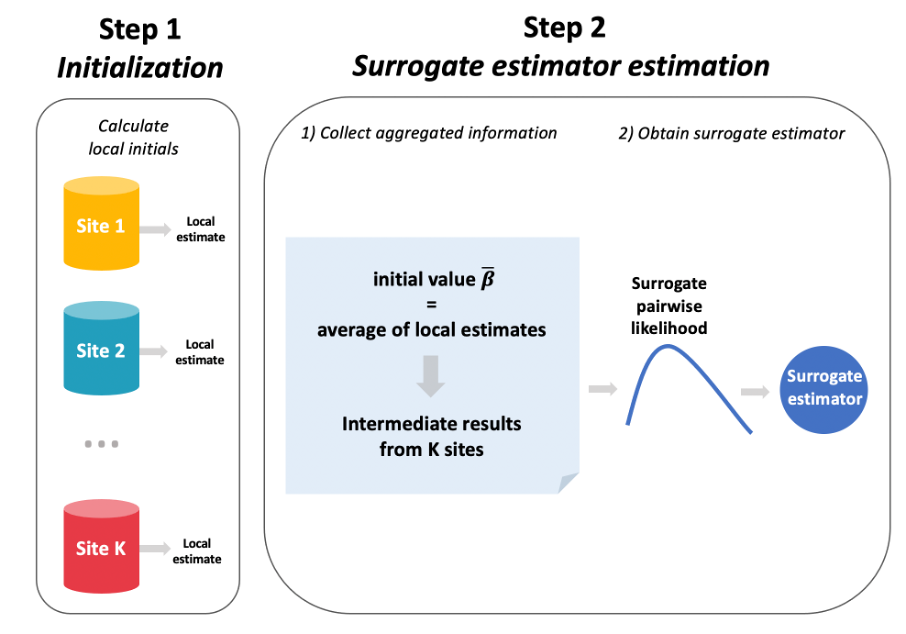
- dCLR workflow
Integrating real-world data (RWD) from several clinical sites offers great opportunities to improve estimation with a more general population compared to analyses based on a single clinical site. However, sharing patient-level data across sites is practically challenging due to concerns about maintaining patient privacy. We develop a distributed algorithm to integrate heterogeneous RWD from multiple clinical sites without sharing patient-level data. The proposed distributed conditional logistic regression (dCLR) algorithm can effectively account for between-site heterogeneity and requires only one round of communication. Our simulation study and data application with the data of 14,215 COVID-19 patients from 230 clinical sites in the UnitedHealth Group Clinical Research Database demonstrate that the proposed distributed algorithm provides an estimator that is robust to heterogeneity in event rates when efficiently integrating data from multiple clinical sites. Our algorithm is therefore a practical alternative to both meta-analysis and existing distributed algorithms for modeling heterogeneous multi-site binary outcomes.