Are you beset by insoluble difficulties?
Do you suffer from a stained conscience, or reputation?
Have immovable objects nullified the unstoppable force that is you?
With new Alkahest-brand solvent, worry no more! Simply apply to the problem at hand, and watch it melt away before your eyes.
Alkahest is effective against stubborn rock formations:

|
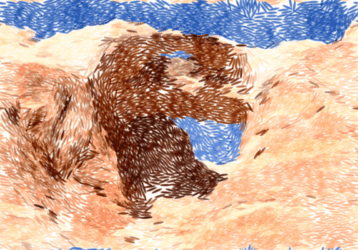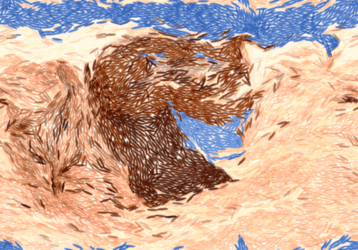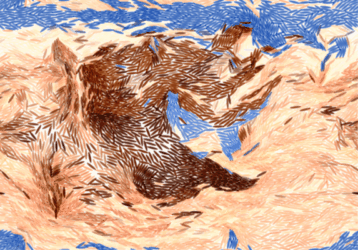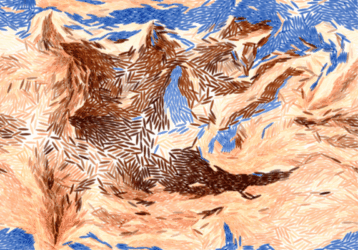
|
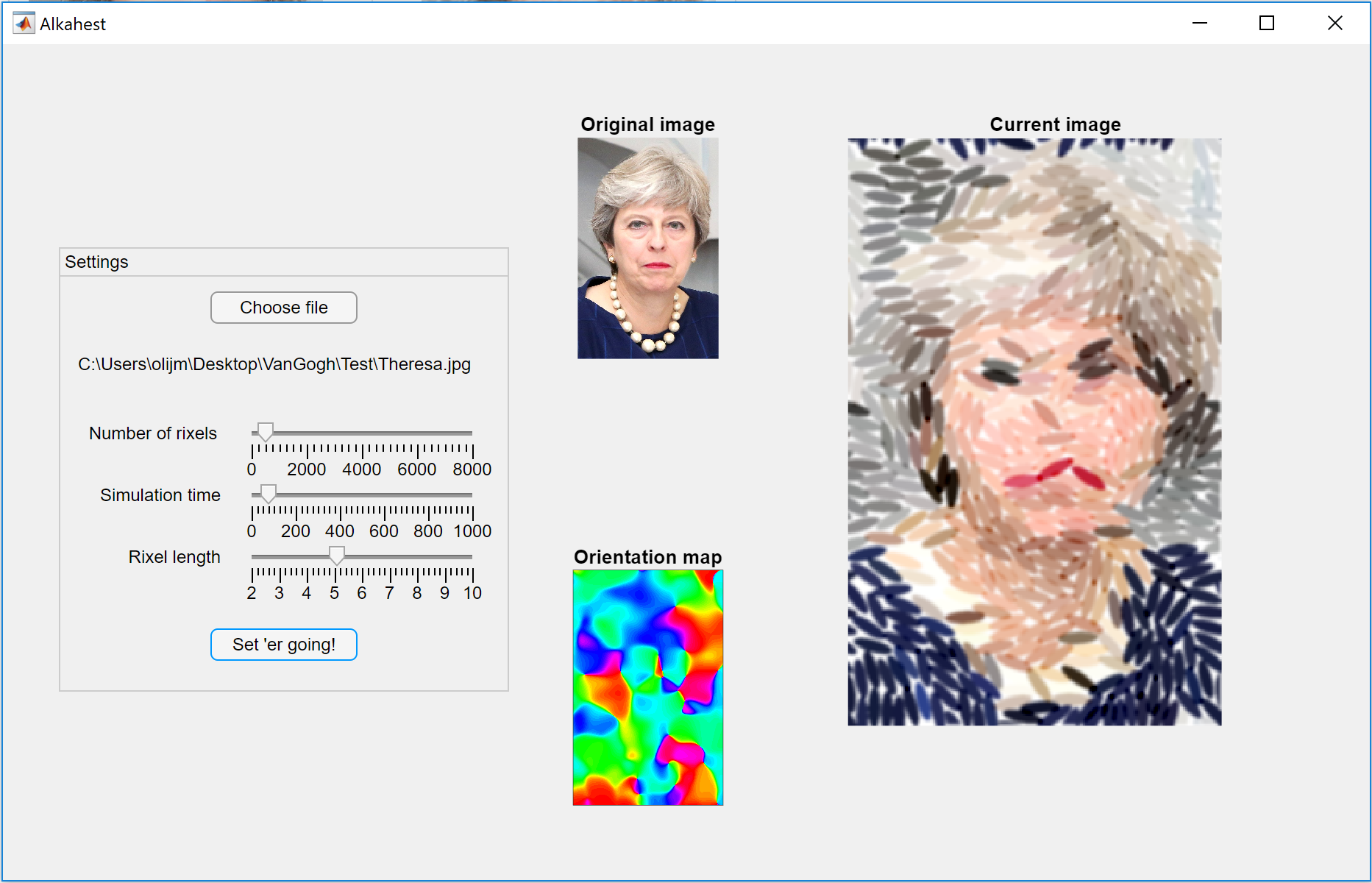
Hard-to-remove Prime Ministers:

|

|
The abstract concept of love:

|

|
And much more. So try it today!
To install Alkahest, simply download and run Alkahest.mlappinstall. This will install it in your copy of Matlab.
Alkahest has been tested on Matlab 2018a and 2018b. If you test it on a another version of Matlab, I would be most grateful if you could report the outcome through the Issues tab.
Finding the target of your ire is dissolving more slowly than you'd like? Alkahest includes a .c file for computing the potential gradients acting on rods (the most time-consuming stage of calculating the dynamics of the system). To use this, simply follow this guide to compiling .mex files, applying it to mexCalcEnergyGradientsPeriodic.c. Alkahest will then automatically detect the compiled version and use it in place of the native .m function.
To begin Alkahest, click on the Alkahest logo in the 'APPS' tab in your main Matlab window. This will bring up the Alkahest GUI:
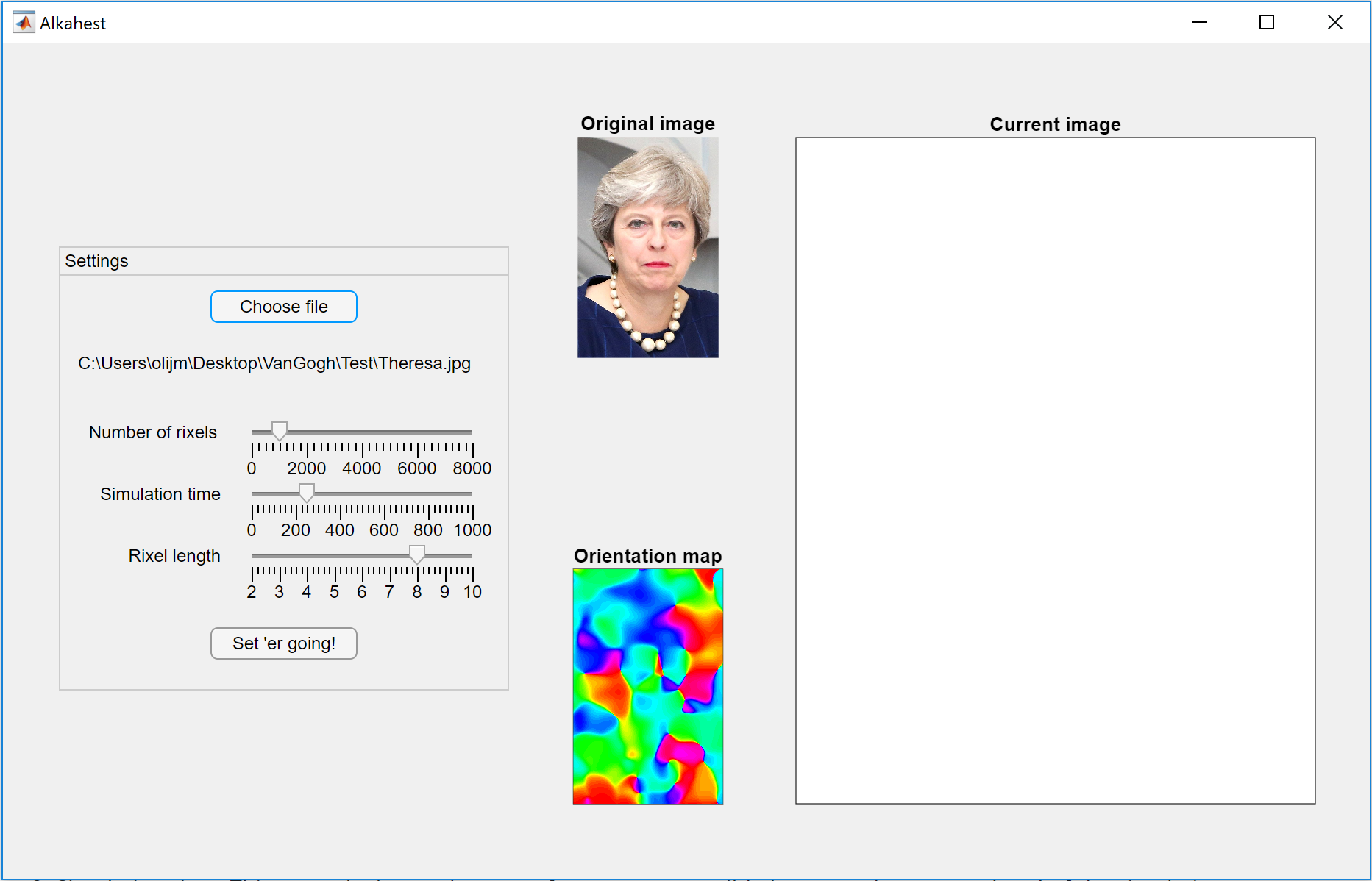
You next need to select the input image. To do this, simply click 'Choose file'. This will bring up a folder selection dialogue. Navigate to the folder containing your image, click 'Select folder', then in the next dialog select your target image. Alkahest will then show the image in the top left axes, and its estimation of the local orientation for each pixel in the image in the botton left axes:
Alkahest uses the tensor method to estimate the local orientation of the image at each location. For more details, see this book (chapter PDF available here).
The Alkahest GUI limits users to three variable parameters, selectable by the sliders within the 'Settings' panel:
- Number of rod-pixels, or 'rixels'. These are the elongated image elements that will actually move during the simulation steps. Much like in normal images, increasing the number of rixels will increase the resolution and clarity of the output images. However, processing times increase substantially at high rixel numbers. Choose wisely.
- Simulation time. This controls the total extent of movement possible between the start and end of the simulation.
- Rixel length. This controls the aspect ratio (length to width ratio) of each rixel. Higher values produce more elongated image elements, and also tend to produce larger-scale motions in the system dynamics.
Once you are happy with your parameter choices, press 'Set 'er going!' to begin simulating the system dynamics. You will see a series of five loading bars appear, with the first four corresponding to the simulation initialisation stages, and the last corresponding to the main simulation.
Depending on the settings chosen, now is probably a good time to go to sleep (while irresponsibly leaving your computer running overnight).
As Alkahest runs, the latest output image will be shown in the large axes on the right of the figure:
Each image is also saved as a .tif in a newly created folder called 'MeltingTimelapse' in the directory your image is saved in. To watch your target melt in real time, you can load these images as a movie using File -> Import -> Image Sequence in Fiji.
- Wensink, H. H., & Löwen, H. (2012). Emergent states in dense systems of active rods: From swarming to turbulence. Journal of Physics Condensed Matter, 24(46). https://doi.org/10.1088/0953-8984/24/46/464130
- Lowen, H., Dunkel, J., Heidenreich, S., Goldstein, R. E., Yeomans, J. M., Wensink, H. H., & Drescher, K. (2012). Meso-scale turbulence in living fluids. Proceedings of the National Academy of Sciences, 109(36), 14308–14313. https://doi.org/10.1073/pnas.1202032109
- Püspöki, Z., Storath, M., Sage, D., & Unser, M. (2016). Transforms and operators for directional bioimage analysis: A survey. Advances in Anatomy Embryology and Cell Biology, 219, 69–93. https://doi.org/10.1007/978-3-319-28549-8_3
- By EU2017EE Estonian Presidency - Tallinn Digital Summit. Welcome dinner hosted by HE Donald Tusk. Tour de table, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=62856180
- By Pretzelpaws at the English language Wikipedia, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=7103158
- Public Domain, https://commons.wikimedia.org/w/index.php?curid=2106343
- Oliver J. Meacock
- William P. J. Smith