-
-
Notifications
You must be signed in to change notification settings - Fork 71
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Create chemical reactions as Network of complexes.md
A network dynamics approach to chemical reaction networks
- Loading branch information
1 parent
336d9e2
commit 87f56c3
Showing
1 changed file
with
146 additions
and
0 deletions.
There are no files selected for viewing
146 changes: 146 additions & 0 deletions
146
docs/src/tutorials/chemical reactions as Network of complexes.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,146 @@ | ||
| Based on https://github.com/SciML/Catalyst.jl/issues/318 and explanation from the paper | ||
| "A network dynamics approach to chemical reaction networks(CRN)" https://arxiv.org/pdf/1502.02247.pdf | ||
|
|
||
| The following is explanation of alternative way to express general chemical reaction networks using a network approach.(a network of intermediate complexes.) | ||
|
|
||
| Imagine a chemical reaction network with `ns` distinct chemical species and has `nr` unidirectional reactions denoted as `r` | ||
| ```julia | ||
| using Catalyst, ModelingToolkit | ||
| using LinearAlgebra | ||
| rn = @reaction_network begin | ||
| k₁, 2A --> B | ||
| k₂, A --> C | ||
| k₃, C --> D | ||
| k₄, B + D --> E | ||
| end k₁ k₂ k₃ k₄ | ||
| r = reactions(rn); | ||
| # Num. of reactions & Num. of species | ||
| nr = numreactions(rn); ns = numspecies(rn); | ||
| ``` | ||
| Let `nc` be the total number of complexes involved in the reaction network, we can easily create an array of `complexes_mat` representing unique complexes involved. | ||
|
|
||
| ```julia | ||
| complexes_mat = []; | ||
| for i ∈ 1:numreactions(rn) | ||
| push!(complexes_mat, [r[i].substrates r[i].substoich]) | ||
| push!(complexes_mat, [r[i].products r[i].prodstoich]) | ||
| end | ||
| complexes_mat = unique(complexes_mat) | ||
| nc = length(complexes_mat) # Number of unique complexes | ||
| ``` | ||
| Since each of the `nc` complexes is a combination of the `ns` chemical species we define the `ns x nc` matrix Z with positive integer elements expressing the composition of the complexes in terms of the chemical species. The k-th column of Z denotes the composition of the k-th complex, and the matrix Z is called the complex composition matrix. Lets create this matrix from the unique complexes set that we created in `complexes_mat` | ||
| ```julia | ||
| # complex composition matrix/complex stoichiometeric matrix | ||
| smap = speciesmap(rn) | ||
| Z = zeros(Int, ns, nc); | ||
| for i ∈ 1:nc | ||
| for j in 1:length(complexes_mat[i][:,1]) | ||
| Z[smap[complexes_mat[i][:,1][j]] ,i] = complexes_mat[i][:,2][j] | ||
| end | ||
| end | ||
| ``` | ||
| Based on previously created `complexes_mat`, it results in a directed graph `G` with `nc` vertices and `nr` edges is called the graph of complexes(have not created thisyet) , and is defined by its `nc × nr` incidence matrix B. | ||
| which is defined as `B` | ||
| ``` | ||
| Bᵢⱼ = -1, if the ith complex is the substrate of the jth reaction, | ||
| = 1, if the ith complex is the product of the jth reaction, | ||
| = 0, otherwise | ||
| ``` | ||
| ```julia | ||
| B = zeros(Int, nc, nr); # Incidence matrix | ||
| for i ∈ 1:nr | ||
| for j ∈ 1:nc | ||
| if isequal([r[i].substrates r[i].substoich], complexes_mat[j]) | ||
| B[j, i] = -1; # found the reactant as complexes | ||
| continue; | ||
| end | ||
| if isequal([r[i].products r[i].prodstoich], complexes_mat[j]) | ||
| B[j, i] = 1; # found the product as complexes | ||
| continue; | ||
| end | ||
| end | ||
| end | ||
| ``` | ||
| It can be immediately verified that `Z.B` equals the standard net-stoichiometric matrix, and this can be verified from catalyst.jl API function `netstoichmat` | ||
| ```julia | ||
| nmat = netstoichmat(rn)'; # in-built API from Catalyst | ||
| new_stoich = Z*B | ||
| isequal(nmat, new_stoich) | ||
| ``` | ||
| It is evident that since `netstoichmat` can be factorized as `Z*B` , we can express the ODESystem represented by reaction network as | ||
| `ẋ = netstoichmat(rn)*v(x) = Z*B*v(x)` | ||
| here, `v(x)` is `oderatelaw.(equations(rn))` i.e. a vector of reaction-rate of reaction system. | ||
|
|
||
| Let `Zₛᵢ` be the column of Z corresponding to the substrate `Sᵢ` of the i'th reaction. | ||
| The reaction-rate of the i'th unidirectional reaction between the substrate `Sᵢ` and product `Pᵢ`, | ||
| `vᵢ[x] = dᵢ[x] kᵢ exp(Zᵀₛᵢ .log(x))` | ||
| where `kᵢ ` is rate-constants , `dᵢ[x]` is a rational function for enzyme kinetics and is `1` for mass-action kinetics | ||
|
|
||
| Lets define an outgoing matrix `Δ` as, | ||
|
|
||
| ``` | ||
| Δᵢⱼ = 0 , if Bᵢⱼ = 1 | ||
| = Bᵢⱼ , otherwise | ||
| ``` | ||
| ```julia | ||
| ## Outgoing matrix | ||
| Δ = copy(B) | ||
| Δ[Δ.==1] .= 0 | ||
| ``` | ||
| It can be shown that shown the `nc × nc` weighted Laplacian matrix associated with the complex graph of | ||
| the network is `L ( x ) = B .KD (x) ∆ᵀ` ,where `K = diagonal_matrix(parameters(rn))` and `D(x) = diagonal_matrix(dᵢ[x])` | ||
|
|
||
| Since our system is mass-action kinetics, we can see that `D(x) = I(nr)` i.e. identity matrix. | ||
| ` K` and `D (x)` are symbolic diagonal matrices ,that are created if we use `LinearAlgebra.diagm` function from julia on `parameters(rn)`, as follows | ||
| ```julia | ||
| ## the Laplacian matrix/weighted Laplacian matrix of the graph of complexes | ||
| # L(x) = B.K.D(x).Δᵀ | ||
| # where K.D(x) = diag([parameters]) of nr x nr size | ||
|
|
||
| rate_consts = parameters(rn) | ||
| KD = diagm((map(Num, rate_consts))) | ||
|
|
||
| ``` | ||
| Now, we are in position to define a weighted laplacian matrix as follows | ||
| ```julia | ||
| L = (B*KD)*Δ' | ||
| dxdt = -Z*L*exp.(Z'*log.(states(rn))) # array of equations | ||
| ``` | ||
| We obtain `dxdt` , as array of equations that we could also obtain by converting `rn` to `ODESystem` with `combinatoric_ratelaw = false` | ||
| Plotting the directed graph `G`, having `complexes_mat` as nodes and arrows/edges as reactions | ||
| `create_graph function` is taken from [YouTube video by Chris Lam](https://www.youtube.com/watch?v=cLl611S4Qe0) | ||
| ```julia | ||
| using LightGraphs,GraphRecipes | ||
| function create_graph(incidence::Array{Int,2}) | ||
| n=size(incidence)[2] | ||
| graph=DiGraph(n) # find nodes where edge goes out and edge goes | ||
| v_out=findall(x -> x ==-1, incidence) # edge goes out of vertex | ||
| v_in=findall(x -> x ==1, incidence) # edge goes in of vertex | ||
| # match the Cartesian index between the two lists, add edge | ||
| for i in v_in | ||
| for o in v_out | ||
| if o[1]==i[1] | ||
| add_edge!(graph, o[2], i[2]) | ||
| end | ||
| end | ||
| end | ||
| return graph | ||
| end | ||
| graph = create_graph(Array(B')) | ||
|
|
||
| complexes_names = [] | ||
| for i ∈ 1:nc | ||
| push!(complexes_names ,sum(complexes_mat[i][:,1].*complexes_mat[i][:,2])) | ||
| end | ||
|
|
||
| graphplot(graph,names=string.(complexes_names), | ||
| nodesize=0.1,nodeshape=:circle, | ||
| curves = false) | ||
| ``` | ||
| 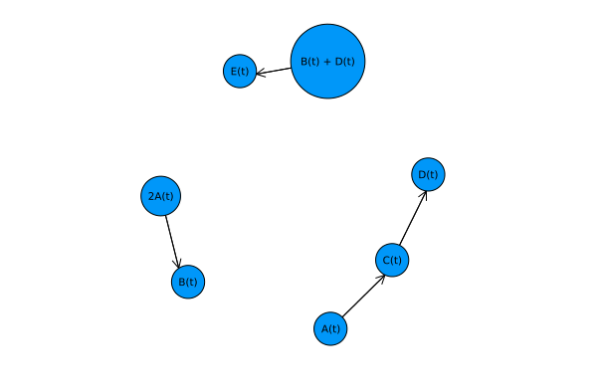 | ||
|
|
||
| Compared to what `Catalyst.Graph(rn)` yields as | ||
| 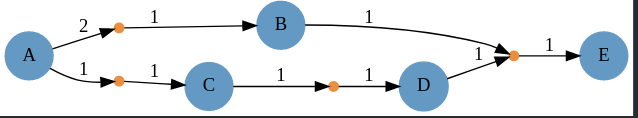 | ||
|
|
||
| **What remains to be done is** | ||
| - 1 Testing this approach for enzyme kinetics and even more kinetics |