The World Health Organization (WHO) characterized the COVID-19, caused by the SARS-CoV-2, as a pandemic on March 11, while the exponential increase in the number of cases was risking to overwhelm health systems around the world with a demand for ICU beds far above the existing capacity, with regions of Italy being prominent examples.
Brazil recorded the first case of SARS-CoV-2 on February 26, and the virus transmission evolved from imported cases only, to local and finally community transmission very rapidly, with the federal government declaring nationwide community transmission on March 20.
Until March 27, the state of São Paulo had recorded 1,223 confirmed cases of COVID-19, with 68 related deaths, while the county of São Paulo, with a population of approximately 12 million people and where Hospital Israelita Albert Einstein is located, had 477 confirmed cases and 30 associated death, as of March 23. Both the state and the county of São Paulo decided to establish quarantine and social distancing measures, that will be enforced at least until early April, in an effort to slow the virus spread.
One of the motivations for this challenge is the fact that in the context of an overwhelmed health system with the possible limitation to perform tests for the detection of SARS-CoV-2, testing every case would be impractical and tests results could be delayed even if only a target subpopulation would be tested.
This dataset contains anonymized data from patients seen at the Hospital Israelita Albert Einstein, at São Paulo, Brazil, and who had samples collected to perform the SARS-CoV-2 RT-PCR and additional laboratory tests during a visit to the hospital.
All data were anonymized following the best international practices and recommendations. All clinical data were standardized to have a mean of zero and a unit standard deviation.
- Hematocrit,Leukocytes,Potassium etc.
- SARS-Cov-2 exam result - target; 1 for positive and 0 for negative test
SMOTE
this technique, minority class data are synthetically over-sampled, presenting for the training subset the same proportion of instances for the positive and the negative class.
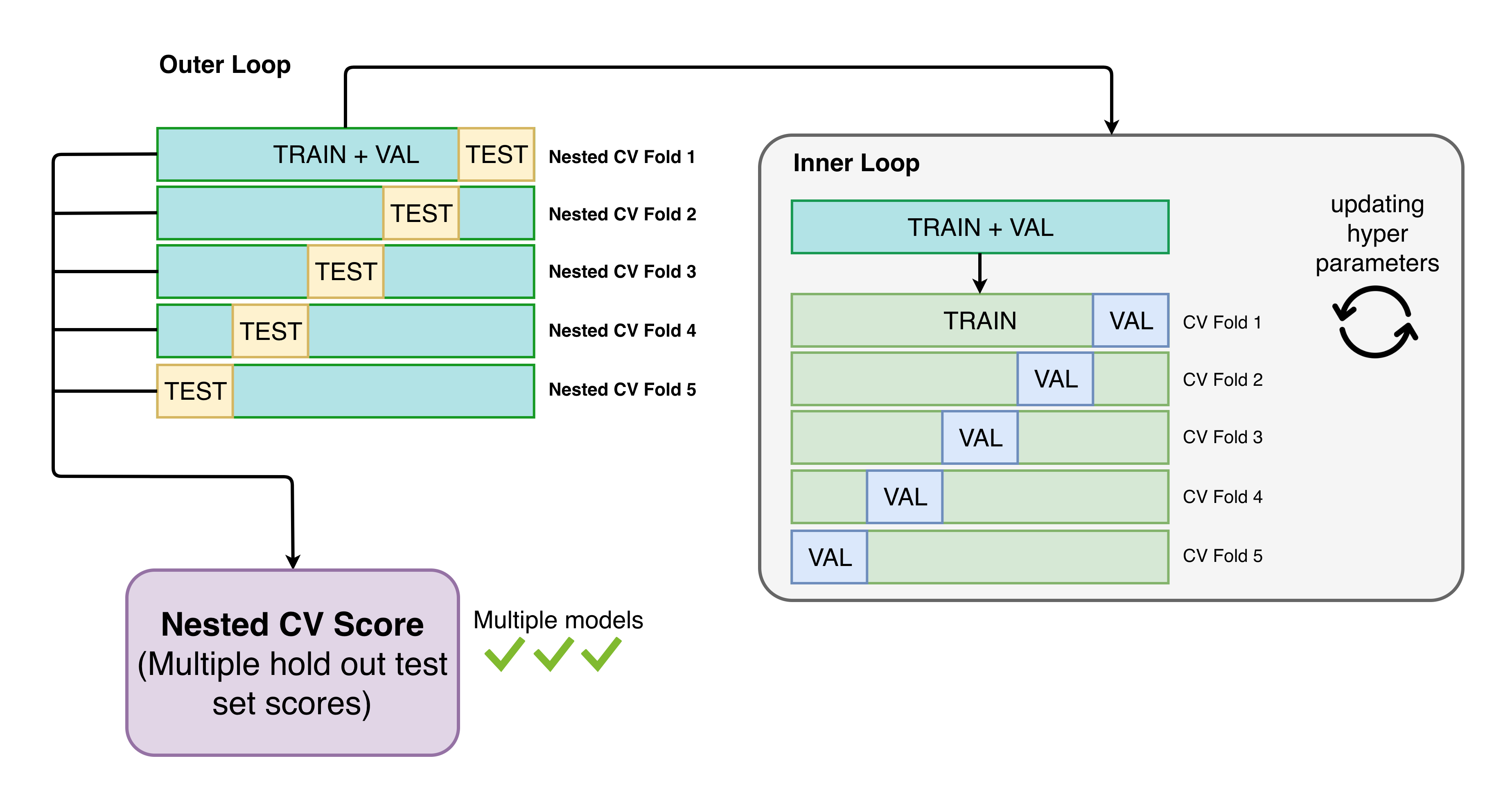
Let us try the following models:
- Random Forest
- XGBoost
- Logistic Regression
Scoring function will be F1, since it is more costly to have false negatives than false positives
- Python 3.6
- Git 2.26
- PyCharm IDEA (recommend)
You can modify or contribute to this project by following the steps below:
1. Clone the repository
-
Open terminal ( Ctrl + Alt + T )
-
Clone to a location on your machine.
# Clone the repository
$> git clone https://github.com/serfati/covid_bloodtest.git
# Navigate to the directory
$> cd covid_bloodtest2. Install Dependencies
# install with pip/conda
$> pip install -r requirments.txt3. launch of the project
# Run nootebook
$> jupyter notebook AML.ipynbauthor Serfati
This program is free software: you can redistribute it and/or modify it under the terms of the MIT LICENSE as published by the Free Software Foundation.