Documentation
TerraFERMA provides a hierarchical options tree and front-end that allows the user to rapidly construct descriptions of multi-physics problems using finite elements. Starting with a blank options file however can be a daunting task. Here we attempt to give a brief overview of the syntax of TerraFERMA by providing a glossary of key terms. This can be useful to prepare new users for what they can expect when looking through the various options. However, the best way to get to grips with TerraFERMA is by working through some of the worked examples, which we describe first in the cookbook. A more comprehsive listing and description of all the options available is the provided before a brief discussion of the overall code structure and development, which is aimed at any developers or debuggers.
The best way to understand TerraFERMA is to try using it. Please download and install the code and then try some worked examples from the cookbook. This is available (and citeable) through figshare:
- TerraFERMA cookbook (version 1.0) doi:10.6084/m9.figshare.1466786
and in the tutorials directory of the source code:
git clone https://github.com/TerraFERMA/TerraFERMA.git
along with worked example input option files. Within the tutorials/manual directory, the cookbook can be built using the command:
cmake -P build.cmake
TerraFERMA adds a layer of control and guidance to the underlying libraries using a hierarchical options tree. This is documented within the front-end GUI, diamond, and also below. Here we provide a short glossary of the key components of TerraFERMA and its core dependencies.
The system is the key unit of a TerraFERMA problem. It collects together sets of coupled fields with nonlinear solvers that contain forms describing the weak-forms of the equations to be solved for the fields. In addition relevant coefficients can be included in a system as well as functionals for diagnostic output and system-wide boundary conditions (e.g. periodic).
Multiple systems can be specified, however, if you want TerraFERMA to solve anything, you will need at least one. Different systems
can "see" fields and coefficients from the other systems however only fields specified within a particular system can be solved for
within the nonlinear solvers of that system. If one system depends on the field values from another system then order matters.
TerraFERMA respects the order in which systems are described in the options file. If two systems are coupled together then it is
possible to iterate between them using a Picard-type scheme by setting options available under /nonlinear_systems. It is generally better to describe
fields that are strongly coupled together within a single system as this allows the use of the better convergence properties of Newton nonlinear solvers.
A system requires a valid (one described under /geometry) mesh name and a unique global ufl symbol be specified.
A field describes a prognostic variable that needs to be solved for. It describes a DOLFIN function and, if multiple fields are described within a single system, is a subfunction of a larger system mixed function.
A field needs a unique global ufl symbol, a rank (scalar, vector or tensor), an element to describe its function space, and an initial condition (if solving a steady problem this is still required as an initial guess for the nonlinear solver). Additionally a field may describe relevant boundary conditions or reference points as well as post-solve modifications like zero points and caps. To be able to interrogate a field in the output it is also necessary to activate diagnostic options, such as visualization, detectors, steady state monitoring and statistics.
A coefficient describes a variable that is either set by the user or determined from other fields and coefficients and does not need to be solved for. It can be either a DOLFIN expression, constant or function type.
All coefficients needs a unique global ufl symbol, a rank (scalar, vector or tensor) and a value. Constant coefficients (of any type) can be simply specified as fixed value floats. Spatially or temporally varying coefficients (of expression or function type) can be specified using either a python or a C++ interface. Using the C++ interface also allows a coefficient to depend on other fields or coefficients.
Coefficients of either expression or function type additionally require an element to be specified. Values at arbitrary points will be interpolated from the nodal values using this element (if you wish to have coefficients evaluated at the integration points then either describe it directly in the forms or use a Quadrature element). Furthermore, if using a function coefficient a full function space will be built for the coefficient and its values stored in a vector. In this case it is necessary for the coefficient to be used in the forms describing the equations or functionals.
As with fields, diagnostics must be activated in order to interrogate the output from coefficients.
A nonlinear solver in TerraFERMA includes both the weak forms of the equations and a description of the method used to solve them. This assumes that the problem is nonlinear but can easily be used for linear problems (which should only require a single nonlinear solver iteration). A nonlinear solver can either be a PETSc SNES Newton solver (most commonly used) or an internal Picard iteration (useful for debugging).
The way the equations are described differs depending on the type of nonlinear solver. For SNES we specify the weak forms of, at least, the residual (linear form) and the Jacobian (bilinear form, normally the automatic derivative of the residual) while for a Picard iteration we describe a linearized set of bilinear and linear weak forms as well as that of the residual (normally just the action of the bilinear form on the solution minus the linear form).
Aside from the equations, all nonlinear solvers require tolerances below which the norm of the nonlinear residual should be
minimized, the maximum number of iterations it may take to do this and a description of the linear solver that
should be used on the bilinear form at each iteration. By default all nonlinear solvers are assembled and solved within
the timeloop (in_timeloop, this exists even for steady problems but only does a single iteration). It is possible to switch this to solve at the
start of the simulation (at_start, useful for steady fields in time dependent problems), only with diagnostic output
(with_diagnostics, useful for fields that
are only needed as diagnostic output) or never (never, mostly used for checkpointing purposes with solvers that were originally solved at
the start). Only nonlinear solvers solved within the timeloop contribute to the global nonlinear residual of all the systems.
Multiple nonlinear solvers can be set per system. This can be useful when solving systems that are challenging to converge and
require some initial solve of approximate equations to improve the behaviour of subsequent solves. If multiple nonlinear solvers
are specified within a system and are solved in the same location (e.g. in_timeloop, at_start, with_diagnostics) then order matters. TerraFERMA respects the order in which solvers are
specified in the options file. Additionally, if multiple solvers are solved in the timeloop then the final one is used in the
global nonlinear residual of all the systems.
The form options allow you to specify the weak form of various components of the equations using the Unified Form Language (UFL) from FEniCS. These can either be linear (e.g. the residual of the equations, which assembles into a vector) or bilinear (e.g. the Jacobian of the residual, which assembles into a matrix).
The forms can use the values of any field or coefficient described anywhere in the options file(hence some care must be taken when assigning ufl symbols to objects) but can only access the test and trial spaces of (and therefore should only describe the solution of) fields from the system which the forms fall under. The form itself also requires a ufl symbol, however this need only be unique within the nonlinear solver that the form belongs to (and not conflict with any of the system, field or coefficient ufl symbols).
Forms are order dependent within a nonlinear solver. Each form within the solver has access to what was
written in the previous forms of that solver. This can be particularly useful if, for instance, you are deriving the Jacobian by
taking the derivative of the residual. In addition all forms can access the UFL specified within global parameters
(/global_parameters/ufl).
A functional is a special class of form that must return a scalar value (i.e. is not tested by any test function). This is used in diagnostic output and any functionals specified are written to the statistics file. Functionals also require a ufl symbol, which need not be unique (except that it should not conflict with those of systems, fields or coefficients).
A ufl symbol is a character or string assigned to a systems, fields and coefficients so that they can be identified within the forms and functionals. In addition, forms and functionals are assigned ufl symbols so that the relevant final calculation can be extracted from the UFL describing the form or functional.
ufl symbols are marked as:
-
globalthese need to be unique across the entire options file
-
solverthese need to be unique within a nonlinear solver and not conflict with any system, field or coefficient ufl symbols
-
functionalthese do not need to be unique but must not conflict with any system, field or coefficient ufl symbols
The values of system, field and coefficient variables can be accessed at different time-levels within forms and functionals using two automatically generated suffixes (TerraFERMA currently uses a two-level time-stepping scheme):
-
<ufl_symbol>_nThe
_nsuffix indicates the previous time-level of the system, field or coefficient with ufl symbol<ufl_symbol>. -
<ufl_symbol>_iThe
_isuffix always points to the latest values the system, field or coefficient with ufl symbol<ufl_symbol>, e.g. within a nonlinear solver<ufl_symbol>_iwill always contain the values of the latest iteration.
In addition to the different time-level discriminating symbols above, system and field ufl symbols are also generated with the following suffixes:
-
<ufl_symbol>_tThe
_tsuffix accesses the test function of the system or field with ufl symbol<ufl_symbol>. These are required when describing any linear form. -
<ufl_symbol>_aThe
_asuffix indicates the trial (or ansatz) function of the system or field with ufl symbol<ufl_symbol>. These may be required when describing a bilinear form. -
<ufl_symbol>_eThe
_esuffix indicates the element of the system or field with ufl symbol<ufl_symbol>. This can be useful when trying to use certain UFL functions.
Bear in mind that for symbols not to conflict with each other they must also not conflict with any of the automatically generated suffixes. It is therefore best to avoid these suffixes when assigning ufl symbols to objects. TerraFERMA performs some preprocessing checks to test for conflicts before building.
A linear solver describes the solver method applied at every iteration of the nonlinear solver. It contains an iterative method and a preconditioner and may optionally contain information about any null spaces that affect the solver.
The iterative method describes a PETSc KSP object for solving linear equations. It accepts any PETSc KSP type but the most commonly used are available through a drop-down list. Most require an error tolerance and the maximum number of iterations in which to achieve that error to be specified. Additionally various "monitors" can be set, that output diagnostic information about the solve to either the log file or other specialized output files.
The preconditioner describes a PETSc PC object for preconditioning the iterative method described
above. It accepts any PETSc PC type but the most commonly used are available through a drop-down list. Some of these should be accessed
through the list as various preconditioner specific options are available. For example the ksp and fieldsplit preconditioners
allow the specification of nested linear solvers, options for which are provided automatically.
The PETSc fieldsplit preconditioner is particularly useful for solving coupled multi-physics problems. Subblocks of the matrix may be
described (using field names, components (for non-scalar fields), region (describing regions of the mesh) or boundary
(describing boundaries of the mesh) ids) and solved separately as a preconditioner for the larger coupled system of equations.
Multiple levels of fieldsplit may be easily described using the hierarchical options system in TerraFERMA.
Various diagnostic outputs are available from simulations run in TerraFERMA. These include statistics and detectors files as well as steady state and convergence monitors. All of these are written to custom data files (derived from the output format used by Fluidity) that can be easily interogated in python or quickly plotted (see additional tools). Visualization output is also available.
All TerraFERMA executables can be run with the command line flags -v<level> and -l. -v controls the level of verbosity and
uses the same levels as DOLFIN from FEniCS. A good starting level (and the default in simulations run through our helper
scripts (see running TerraFERMA and additional tools) is -vINFO, which will print
various information to stdout and stderr about the progress of a simulation. If also run with -l simulations will redirect stdout
and stderr to terraferma.log-<process rank> and terraferma.err-<process rank> respectively. Looking through these log can
provide additional useful diagnostic debugging output. In particular some of the monitors available in the linear
solvers are output here.
The statistics file (<output_base_name>.stat where the output base name is set under /io/output_base_name) contains the
minimum and maximum of all selected fields and coefficients at every statistics dump period
(/io/dump_periods/statistics_period or /io/dump_periods/statistics_period_in_timesteps). In addition selected fields will have the
minimum and maximum of their residuals output. The minimum and maximum are taken as the smallest and largest values of function vectors (fields and
coefficients of type function) or the smallest and largest values of coefficients (of type expression or constant) evaluated at the
mesh vertices. The statistics file is also where all functional values are written by default. The file is easily parseable and
plottable (see additional tools).
The detectors options enable static point evaluations of selected fields and coefficients. Their locations are described under
/io/detectors and the resulting locations and values are output to the detectors file (<output_base_name>.det where the output base name is
set under /io/output_base_name) at every detectors dump period (/io/dump_periods/detectors_period or
/io/dump_periods/detectors_period_in_timesteps). The detectors file is easily parseable and plottable (see additional
tools).
The steady state options enable time-dependent simulations to be monitored for and terminated upon detection of a steady state
(given some tolerance specified under /timestepping/steady_state).
In addition the change in the fields and functionals selected to be monitored for a steady state are output
to the steady state file (<output_base_name>.steady where the output base name is
set under /io/output_base_name) at every steady state dump period (/io/dump_periods/steady_state_period or
/io/dump_periods/steady_state_period_in_timesteps). The steady state file is easily parseable and plottable (see additional
tools).
The convergence of various iterations within TerraFERMA can be monitored in a number of ways. For example PETSc objects like SNES nonlinear
solvers and linear solvers can have their residual and preconditioned residuals monitored,
outputing them to the log file during each solve. If a more parseable format is required then convergence files
(<output_base_name>_....conv where output base name is set under /io/output_base_name and the rest of the file name depends on
the object being monitored but generally includes the system name and the object name and type) can be activated under monitors to have various
convergence monitoring norms output to a parseable and plottable file (for more details on how to parse and plot these files see
additional tools). Outputting convergence files does carry an additional computational expense so should not
generally be done during production runs.
TerraFERMA outputs visualization files in VTK format. The values of fields and
coefficients selected to include in visualization output are included in the visualization files
(<output_base_name>.pvd, which wraps <output_base_name><visualization dump number>.vtu, where output base name is set under
/io/output_base_name) every visualization dump period (/io/dump_periods/visualization_period or
/io/dump_periods/visualization_period_in_timesteps).
Additional visualization output is available to monitor solver convergence. These convergence visualization files will have longer names, identifying the system and solver name and possibly type, and will incur additional computational expense to output.
It is possible to select the function space the visualization is output on under /io/visualization/element
from the short list supported by VTK. This defaults to P1, equivalent of evaluating/interpolating all selected fields and
coefficients to the mesh vertices. If a simulation defines and uses more than one mesh, then separate visualization output files will be generated
for each mesh and the fields and coefficients will be assigned to a mesh based on the system in which they are defined.
We generally use paraview to view the visualization output files.
Buckettools is not an option in TerraFERMA, however you will see the term in a number of places. It arises because the installable library component of TerraFERMA is hierarchical, in a manner reminiscent of the hierarchy of options presented to the user. The highest level object in this hierarchy is known as the bucket, because we throw all the information needed to run the simulation into it. Therefore, the suite of tools, objects and the library itself has become known as buckettools. For more information about the code structure of TerraFERMA (and buckettools) see below.
TerraFERMA inherits most of its functionality from underlying libraries. As such most of the syntax of TerraFERMA is also inherited with some additional terms describing the common interface TerraFERMA imposes. Here we briefly summarize some of this inherited syntax, however, some familiarity with the underlying libraries is always beneficial. Extensive documentation for each of the principal dependencies can be found here:
A Krylov subspace method for solving linear equations. An extensive range of methods are available from PETSc and can be selected under the iterative method for a particular linear solver in TerraFERMA. Generally a KSP is combined with a preconditioning PC.
A preconditioner to be used in combination with a
KSP (or KSP type none). An extensive range of preconditioners is available from PETSc and can be selected under the
preconditioner for a particular linear solver in TerraFERMA. Of particular note is the
fieldsplit preconditioner which is useful for solving coupled multi-physics problems and easily described in the hierarchical
structure of the TerraFERMA options tree.
A scalable nonlinear equations solver, which uses
KSPs to solve linear systems at each nonlinear iteration. A range of SNES types and options are available from PETSc, some
of which are exposed in TerraFERMA when using a SNES type nonlinear solver. Generally a line search (ls)
type is used but a reduced space variational inequality (vi) is useful for constrained systems.
An index set is used to index into PETSc vectors and
matrices. In particular they are used to define arbitrary fieldsplit preconditioners and are referenced in
some of the monitors that are used to debug these structures.
The unified form language is a component of the FEniCS project that allows the high-level description of variational forms for the finite element discretization through a python module. It is used throughout TerraFERMA to write forms and functionals and (behind the scenes) to describe finite element spaces for systems, fields and coefficients.
The FEniCS form compiler is a compiler for variational forms that translates UFL into C++ assembly code. TerraFERMA uses FFC to generate C++ finite assembly code from the description of the equations in the options file. The TerraFERMA library then links to this code to make application (and options file) specific executables.
DOLFIN is the main user interface of FEniCS, additionally providing interfaces to important data structures and finite element assembly algorithms. TerraFERMA uses numerous components of DOLFIN including its mesh data format, wrappers around field and coefficient data structures, boundary condition descriptions and assembly algorithms.
A function is a component of DOLFIN that describes a variable on a function space with an associated vector of values at the degrees of freedom of that function space. TerraFERMA uses functions to implement fields and function coefficients.
An expression is a component of DOLFIN that allows a space dependent variable to be described. It uses an associated finite element to interpolate the values from the degrees of freedom on a cell to arbitrary points within it but does not store a vector of these values, instead evaluating them for each cell. TerraFERMA uses expressions to implement expression coefficients.
A constant (in this context) is a special case of a DOLFIN expression for space invariant variables, which do not require an associated finite element space. TerraFERMA uses DOLFIN constants to implement constant coefficients.
A schema is a rules file built on the SPuD base language that defines the options a model takes and the hierarchical relationship between them. The schema is then used to populate the GUI (diamond) for the model and define valid options input. TerraFERMA uses a schema to describe its hierarchical options tree, which includes documentation visible within the GUI.
Diamond is a GUI provided by SPuD that is automatically populated with an options tree described in the schema. TerraFERMA uses diamond as its front end as well as the front end to some of the additional tools.
The glossary above describes some of the key syntax used in TerraFERMA and its dependencies. However, a typical options file can have many more options than those described above. To help cope with this we have described each option within the TerraFERMA schema so that as a user browses the options tree in diamond, each available option should be briefly explained in the 'description' frame. To make this more easily accesible we also provide these descriptions online in an easier to read and reference format:
While we have tried to document as much as possible as we have developed TerraFERMA, we recognize that some of these descriptions may be short (or non-existant). Please raise any confusing or missing descriptions as an issue on the issue tracker for us to improve.
TerraFERMA principally comprises two components, an installable library and associated tools known as buckettools and an options specific executable that isn't compiled until runtime.
Buckettools is the core of TerraFERMA, containing the vast majority of the code. It is designed, in principle, to be a standalone library. In particular, we aim not to "steal main." This means that the non-buckettools code base of TerraFERMA proper is very lightweight. In fact main.cpp consists of only a few lines:
#include "BucketTools.h"
int main(int argc, char* argv[])
{
buckettools::init(argc, argv);
buckettools::SpudBucket bucket;
bucket.fill();
bucket.run();
return 0;
}
demonstrating how most relevant code lies in buckettools (or our principal dependencies). In principle it should be possible to write a new main that uses buckettools but couples it to another library or model.
The compilable component of buckettools is written in C++ and uses a series of nested classes to mimic the hierarchical options presented to the user:
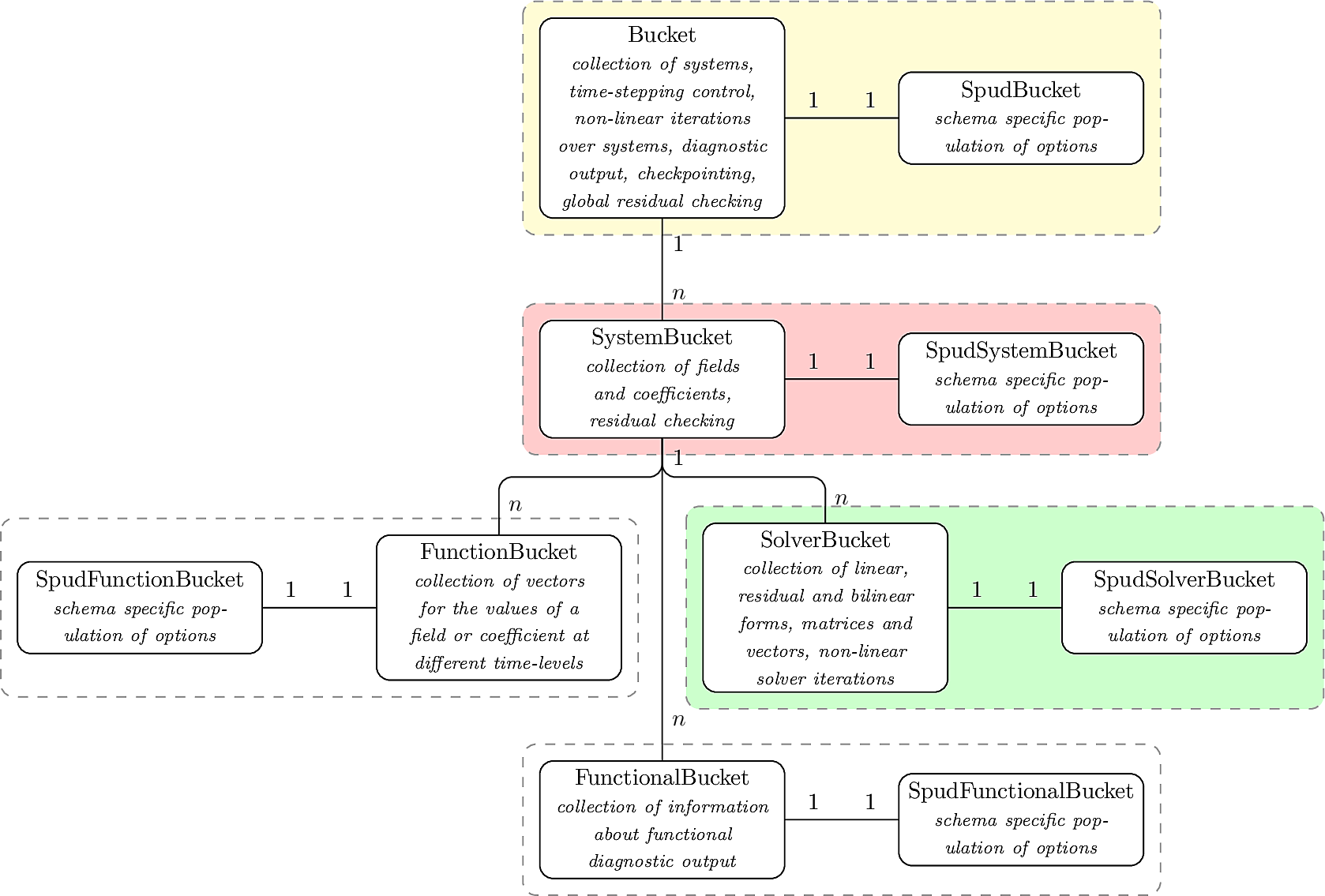
At the top of the hierarchy is the Bucket class, which controls timestepping, checkpointing, global residual monitoring and most diagnostic output. It also contains any number of members of type SystemBucket class, which are equivalent to the system level in the options tree, containing any number of fields and coefficients (FunctionBucket class), nonlinear solvers (SolverBucket class) and functionals (FuntionalBucket class).
Each member of this hierarchy of C++ classes has a corresponding derived class prefixed with Spud (e.g. SpudBucket, SpudSystemBucket etc.). These populate the base class structures based on the TerraFERMA schema and are kept separate to allow models with different schemas to hypothetically reuse the same data structures - envisaging again the possibility of linking buckettools with another model or library and controlling both from the same schema. Such a development strategy would be challenging as large amount of what TerraFERMA/buckettools does is parse user options so these derived classes can be quite large in comparison to the base classes they populate.
This hierarchy of C++ classes forms the backbone of the buckettools library however there are a significant number of other classes and subroutines that are required in buckettools. For example, these include:
-
diagnostic output classes like the statistics file (StatisticsFile), steady state file (SteadyStateFile), detectors file (DetectorsFile), etc., which are all derived from the base diagnostics file class (DiagnosticsFile)
-
detectors classes to store information about the location and value of detectors like PointDetectors and PythonDetectors, which are both derived from GenericDetectors
-
expressions classes, derived from DOLFIN expressions, for initial conditions (InitialConditionExpression), discontinuous expressions over mesh regions (RegionsExpression) and semi-Lagrangian expressions for looking up the values of fields and coefficients at the launch point given some velocity (SemiLagrangianExpression)
The C++ hierarchy is also mimicked in a python buckettools module, which is primarily used for preprocessing the options files, calling ffc, and writing some of the autogenerated code. In addition the module contains some additional tools and post-processing modules (for, for instance, parsing the diagnostic output).
The hierarchical structure of the options tree and code can also be seen in a flowchart describing the series of nested loops comprising TerraFERMA:
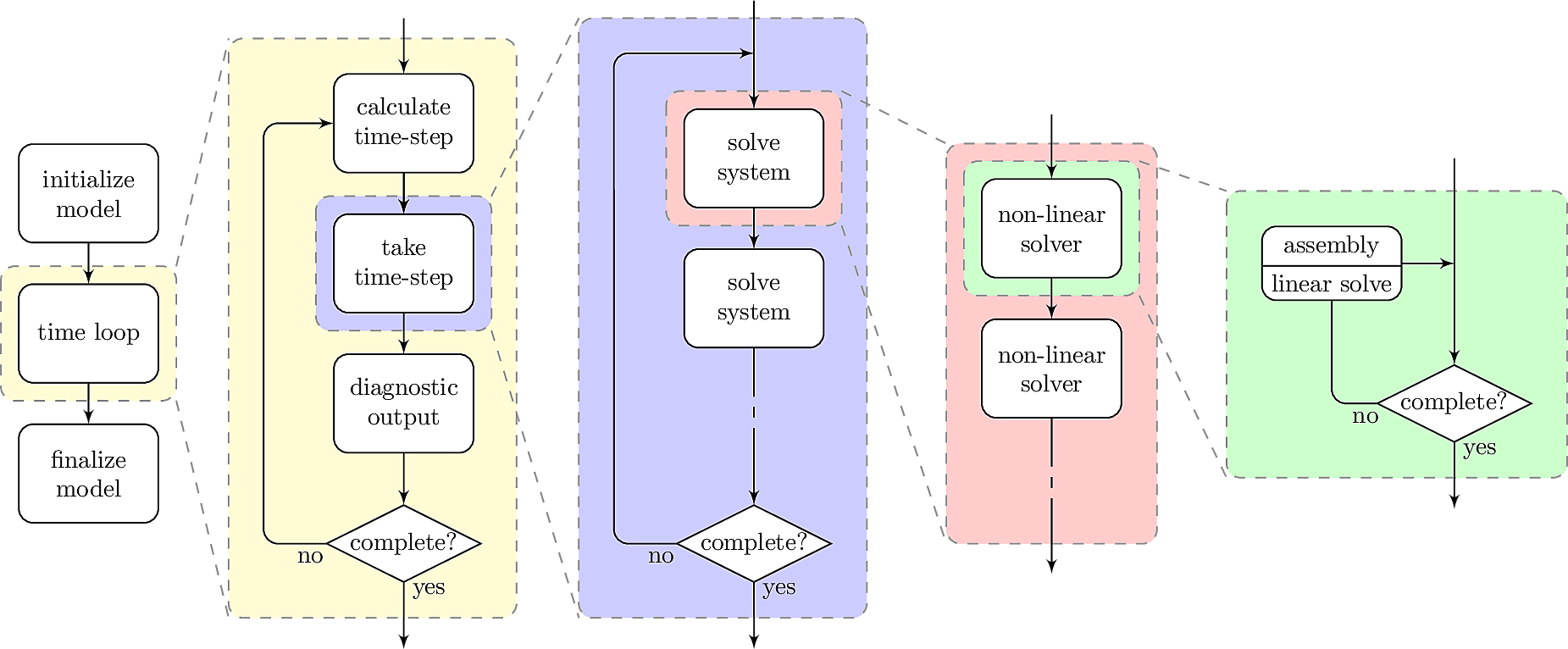
Here the colors correspond to those used in Figure 1 for the different classes of buckettools, roughly delineating which levels each corresponds to at runtime. This is a fairly standard flow diagram for geodynamic models, what we believe makes TerraFERMA more unique is the flexibility included in our set-up.
The development of TerraFERMA (and buckettools) is tied to that of the underlying libraries. In particular we have found ourselves to be particularly dependent on the version of the DOLFIN component of FEniCS. This ends up being obvious in the structure of our development branches in the repository, which roughly follows the tree:
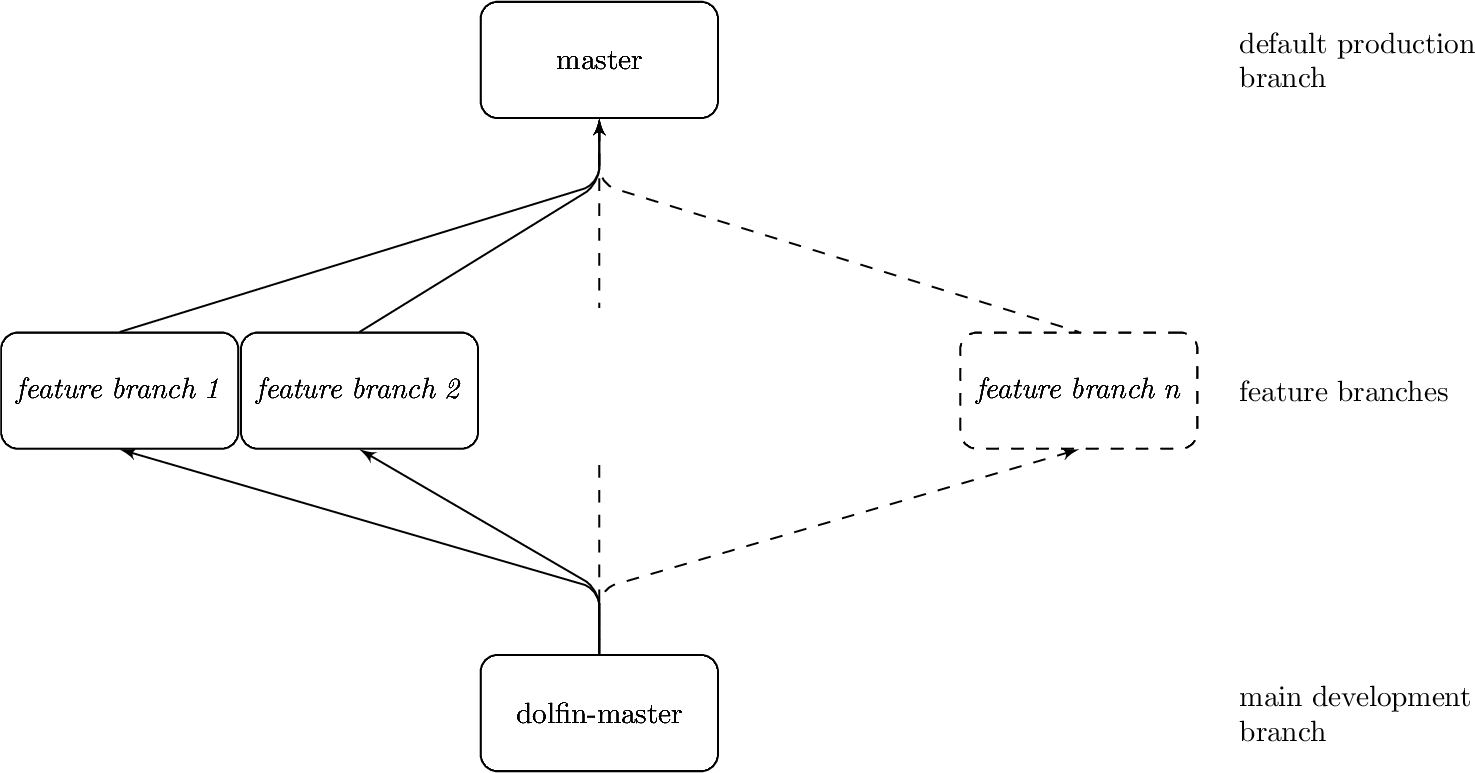
We aim to do the majority of development in a manner consistent with the main branch of DOLFIN (although this normally means the
tferma-master-2019.1.0 branch of our fork of DOLFIN, which lags behind the main DOLFIN repository). This happens in the
2019.1.0/fenics branch of
TerraFERMA. If we need to modify DOLFIN to get some new feature of TerraFERMA to work we will set up a branch off 2019.1.0/fenics
and develop a feature branch, corresponding to a similar feature branch in our fork of DOLFIN (or any other dependency or
component). We do this to more easily propose self-contained pull requests into the main repositories of our dependencies.
This development and feature branch structure means that our main production branch, main, is actually the result of merging
several feature branches, which may still require active maintenance. It also means that the main branch generally does not have any
independent code (other than conflict resolutions) in it. Pull requests of code branched off main will therefore be backported
either into 2019.1.0/fenics or an appropriate feature branch.
It also means that we maintain tferma-master-2019.1.0 branches of the relevant dependencies (see the installation
instructions). At the time of writing this is only required for the FEniCS components. The tferma-master-2019.1.0 branches
similarly consist of merged feature branches in the dependency of interest, with no independent code (other than conflict
resolutions).