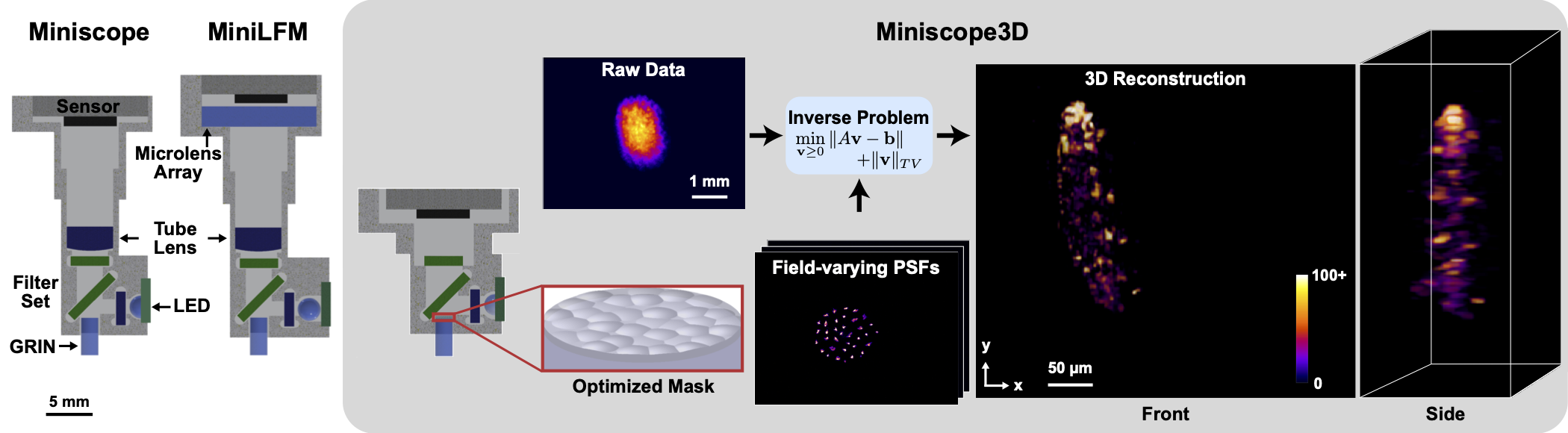
Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy
Please cite the following paper when using this code or data:
Yanny, Kyrollos, et al. "Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy." Light: Science & Applications 9.1 (2020): 1-13.
@article{yanny2020miniscope3d,
title={Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy},
author={Yanny, Kyrollos and Antipa, Nick and Liberti, William and Dehaeck, Sam and Monakhova, Kristina and Liu, Fanglin Linda and Shen, Konlin and Ng, Ren and Waller, Laura},
journal={Light: Science \& Applications},
volume={9},
number={1},
pages={1--13},
year={2020},
publisher={Nature Publishing Group}
}
Sample data and PSFs (needed to run the code) can be found here
This includes the following files:
- calibration.mat - includes the calibratated point spread function, filter function, and wavelength list
- four sample raw measurements
Clone this project using:
git clone https://github.com/Waller-Lab/Miniscope3D.git
First, download the sample data folder mentioned above. This contains PSF components and weights needed for the shift-varying forward model as well as waterbear raw data.
Then, open the Miniscope_3d_shift_varying_main.m file in the 3D Reconstruction folder. This file is the main file that runs the reconstruction. You first need to edit the psf_path variable to the folder path where you downloaded the weights and components (SVD_2_5um_PSF_5um_1_ds4_components.mat and SVD_2_5um_PSF_5um_1_ds4_weights.mat).
Then, run the file, a UI will show up asking you to choose a measurement file and background file. The measurement file is the data you want to deconvolve (e.g. waterbear raw data) and the background is a background image captured before taking the data. This background is subtracted from the data file (to account for back reflections)
This code performs adaptive stitching for randomly spaced microlenses that are multifocal and have aberrations added to them. The adaptive stitching is needed for the microlenses to be correctly 3D printed with Nanoscribe. The code places the stitching artifacts at the boundaries of the microlenses and thus maximizes optical quality.
The demonstration here will be for the 36 multifocal microlenses with astigmatism and tilt added. This is the phase mask used in the paper. The code also provides the necessary .gwl files needed for the print-job on Nanoscribe
This repository contains code for the Miniscope3D paper. The code includes: 1) 3D Reconstruction with Shif-Varying model (matlab). This code reconstructs 3D volumes from a single 2D image. 2) Adaptive Stitching with Nanoscribe (python). This code performs the adaptive stitching needed to 3D print the phase mask with Nanoscribe. We might be adding Microlens phase mask optimization (python) in the future.