New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
How can I add colored tile under the tip label? #25
Comments
|
Also, I can't set the param |
This is because you use
This is because the You can refer to the following codes. |
|
Thank you very much! The problem that had been bothering me for a long time was finally solved. |
I learned that use geom_fruit and geom_tile can add bars to the circular tree graph, but when my tip nodes do not have the same x value, some tiles cannot be shown on the plot with the warning message
and here is my output figure:
It's clear that node "Microcoleus_asticus_IPMA8" has no colored shading.
I want to get a figure like these followings, having colored ring under species the tip label, and grouped by phylum.
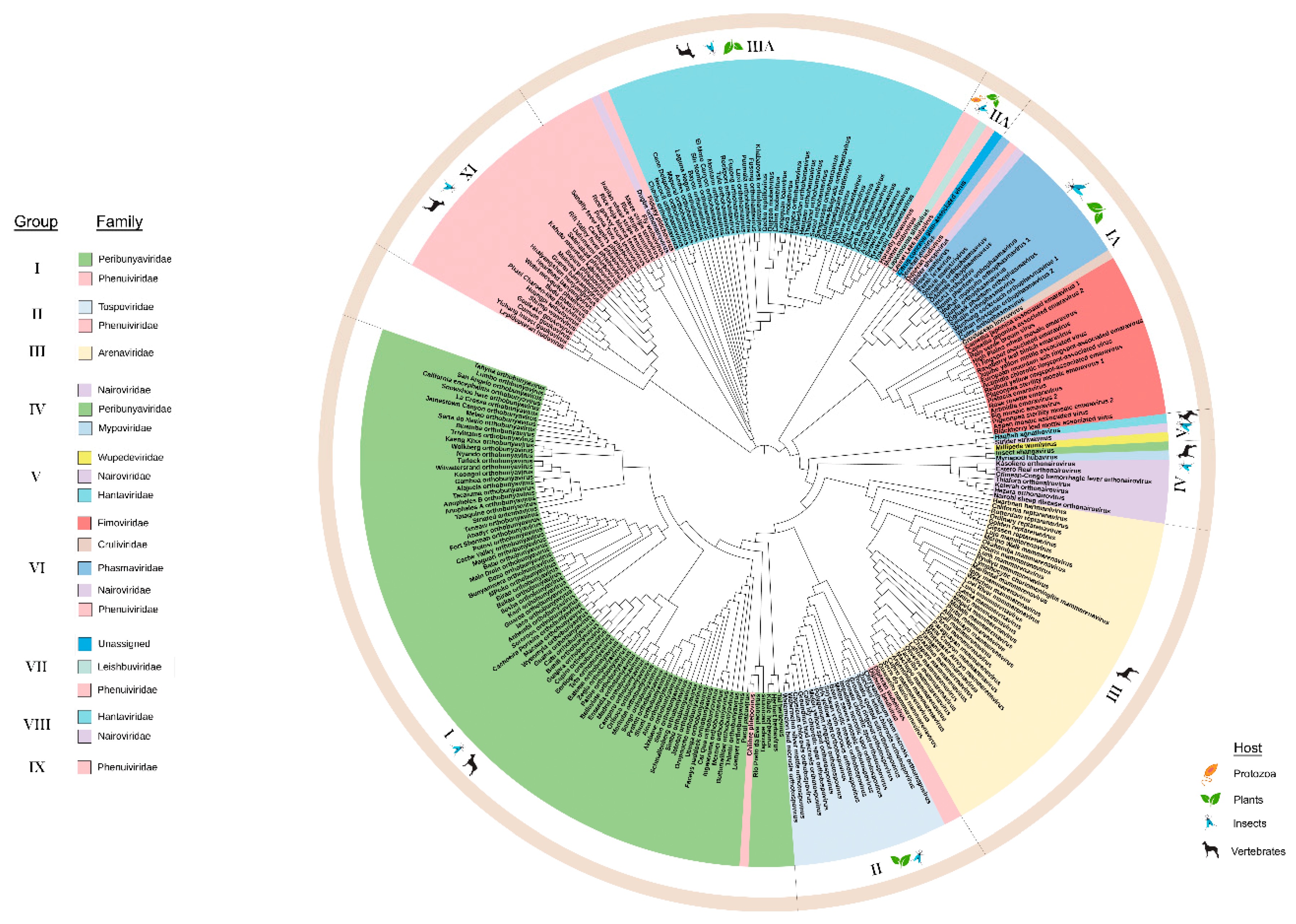

Could you please offer me some help? How can I make figures like these using ggtree and ggtree extra?
Here is my code and origin file:
tree_file:
id50.txt
csv_file:
id50_items.csv
The text was updated successfully, but these errors were encountered: