New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[SYSTEMDS-3393] Implement SIMD usage for basic dense dense MM #1643
Conversation
|
This looks pretty good. I think Alpha has wider SIMD registers, which explains why most configurations perform better in Alpha. |
Note the varying columns case for Alpha though, Lima is faster with Yes, the flag is |
|
What if you use some of the other experimental JVM that should have better support ? |
Which ones do you have in mind, and what kind of support do you expect (which the current does not support)? To clarify, do you expect better performance or that we can remove the |
Project Panama: https://openjdk.java.net/projects/panama/ JDK 17 only officially have the API, this does not guarantee that the instructions are correctly vectorized. Hence i am positively looking at your improvements already/ |
|
I ran the experiments again with jdk-19 (early access). |
a07ccd8
to
b208866
Compare
|
Closed by 9bf0a9f (messed up the commit message) |
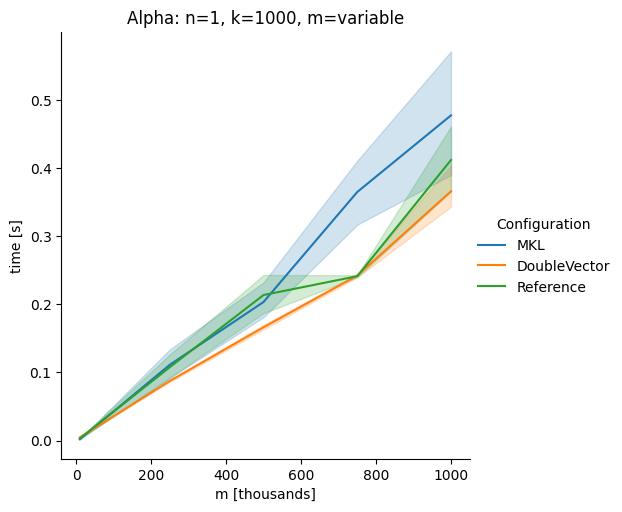
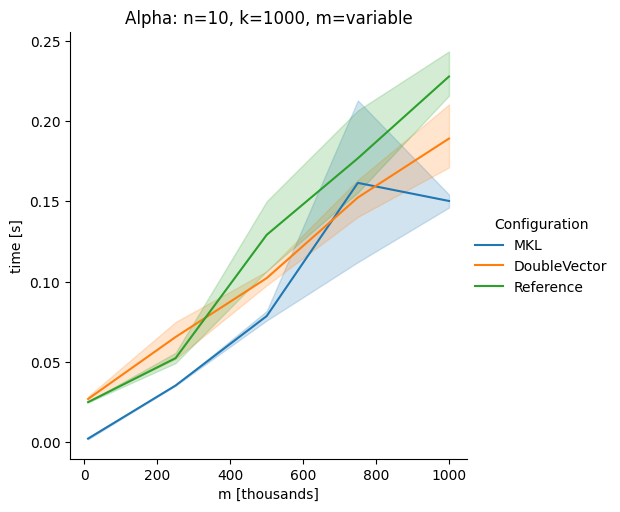
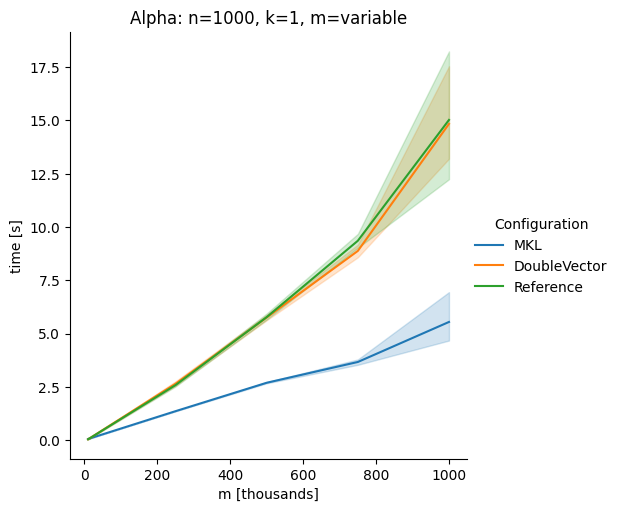
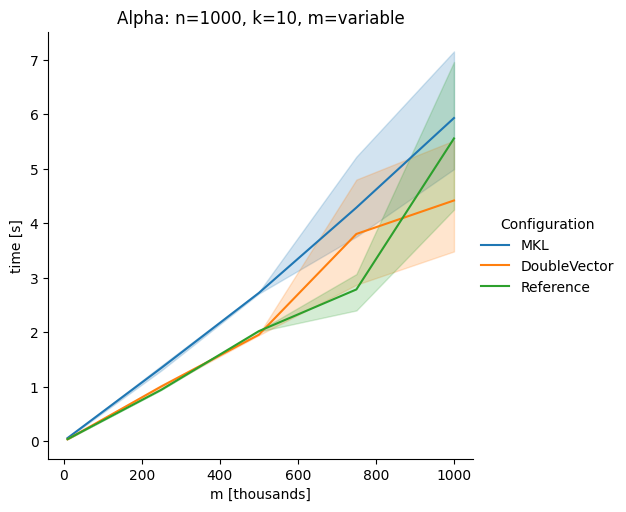
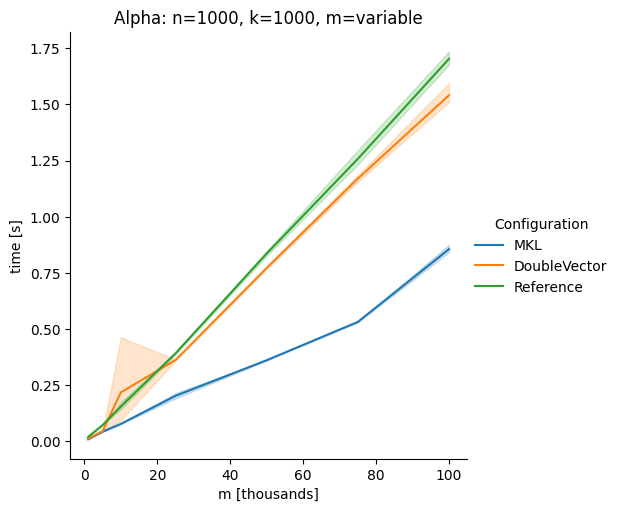
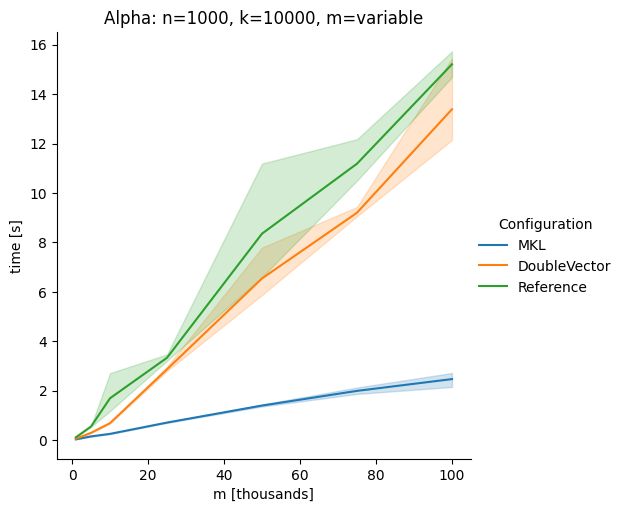
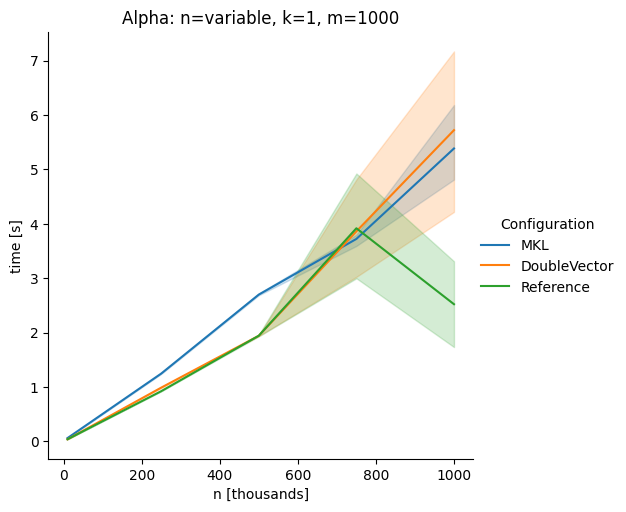
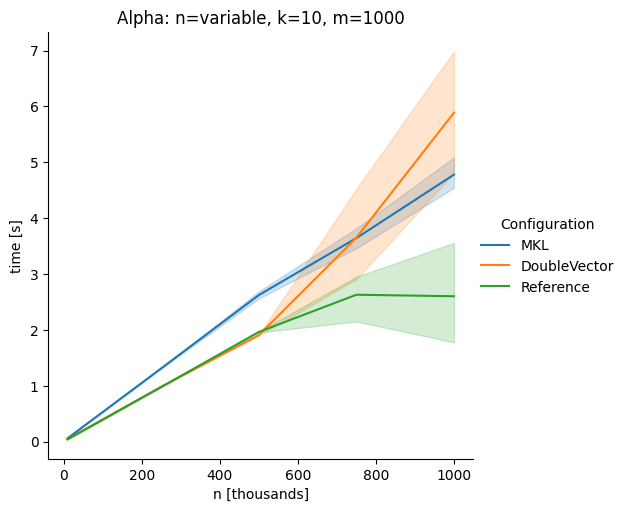

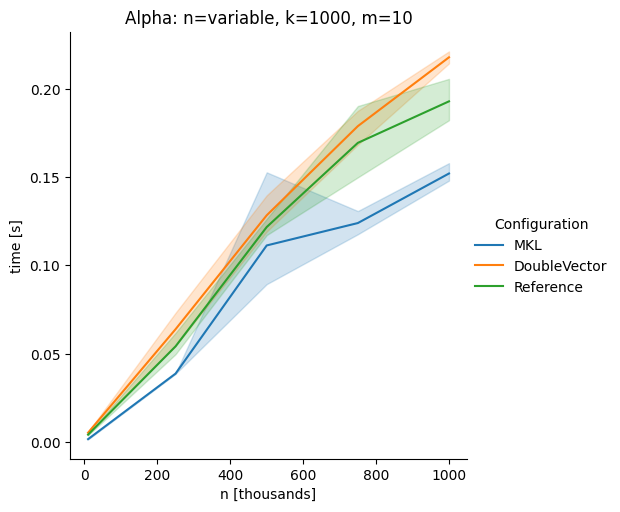
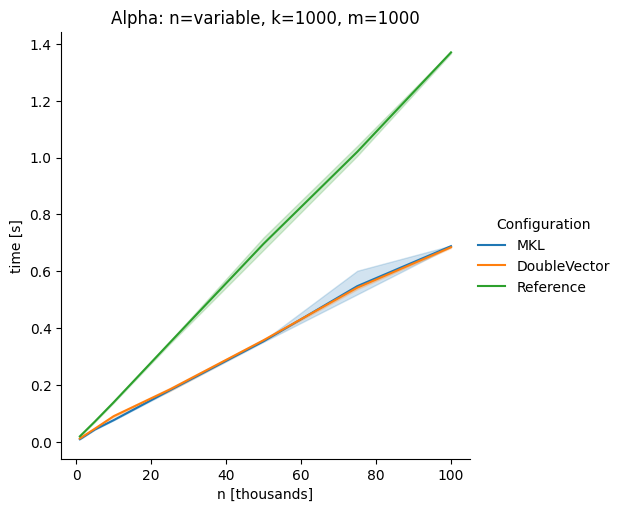
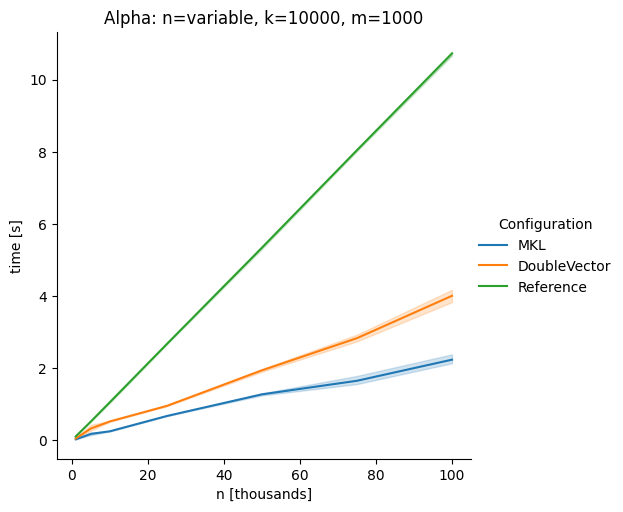

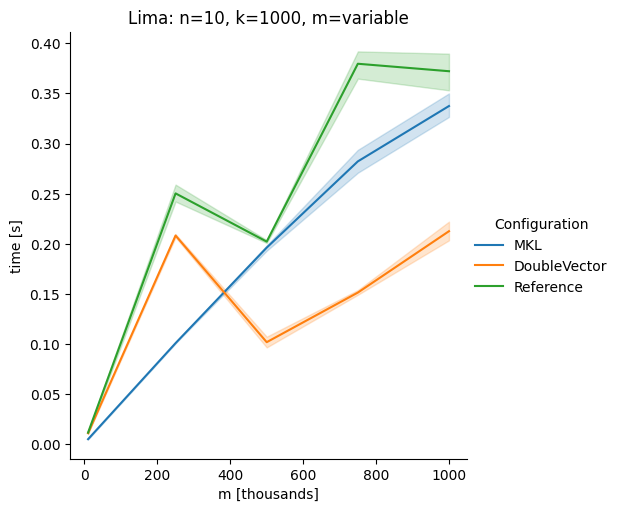

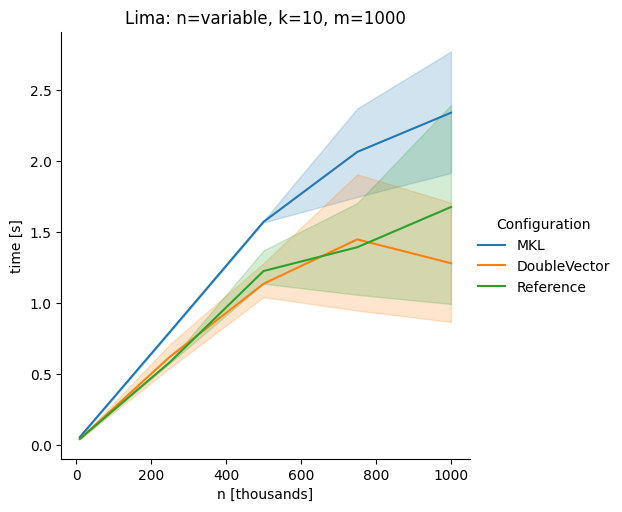



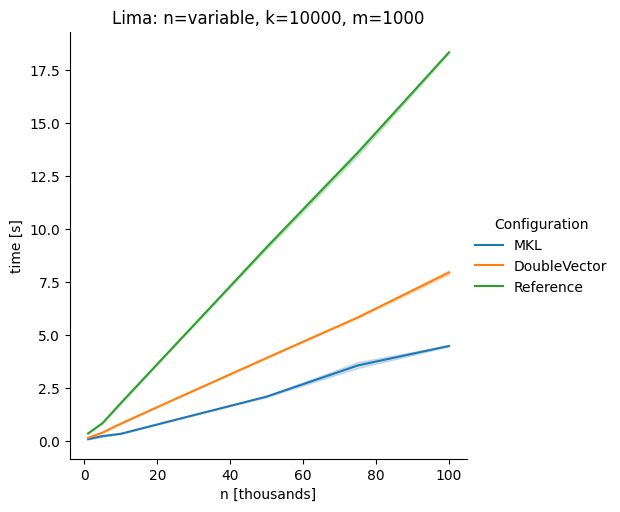
DoubleVectorreplacement for matrix multiplyJDK 17 adds
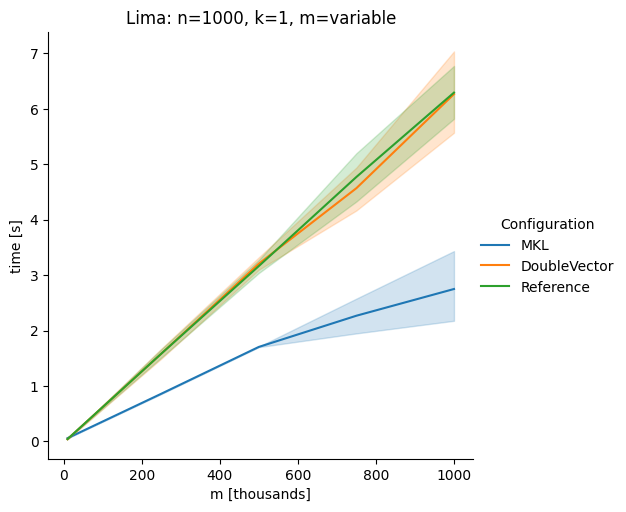
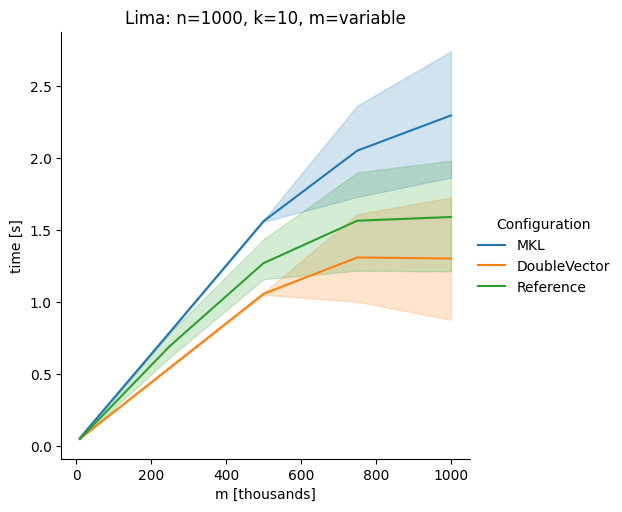
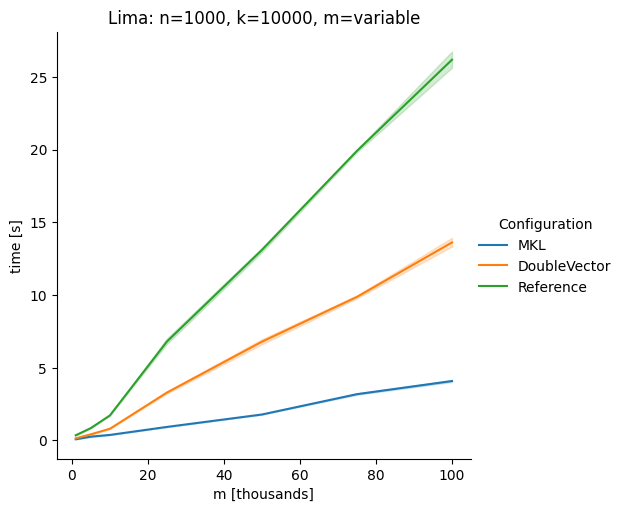
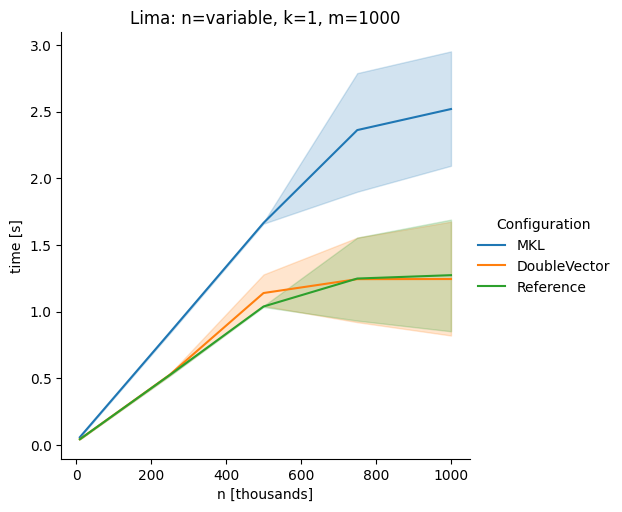
Vectorclasses to use SIMD instructions. This PR replaces the basic dense dense matrix multiply with an equivalentDoubleVectorimplementation. It is necessary to use JDK 17, therefore we should not merge this yet, but keep it in staging for future reference.As an experiment we check a simple matrix multiply:
$X\in \mathbb{R}^{n\times k}, Y\in \mathbb{R}^{k\times m}$
Z = X %*% Y,The experiment script performs 10 matrix multiplications and saves the time of the last 5 to give the JVM some time to optimize.
Vary rows n, m fixed at 1000
Alpha Node
Lima Node
Vary cols m, n fixed at 1000
Alpha Node
Lima Node
Conclusion
The implementation seems to boost the performance in most cases. The case where we vary the number of columns n on the alpha node needs some more exploration, but it seems we are never worse than the current implementation.
Experiment Script