A GUI-based framework for deep-learning enabled segmentation, tracking, time-series analysis of the full Saccharomyces cerevisiae lifecycle
Read the paper Deep learning-driven imaging of cell division and cell growth across an entire eukaryotic life cycle
Developed at the Miranda Laboratory at NCSU
- FIEST Tracking Algorithm: Enhance time-series resolution up to 16x using a generative video interpolation model[1], used to enhance cell-tracking and lineage reconstruction
- Load, analyze, and segment multiple experiments containing numerous phase/flourescent channels
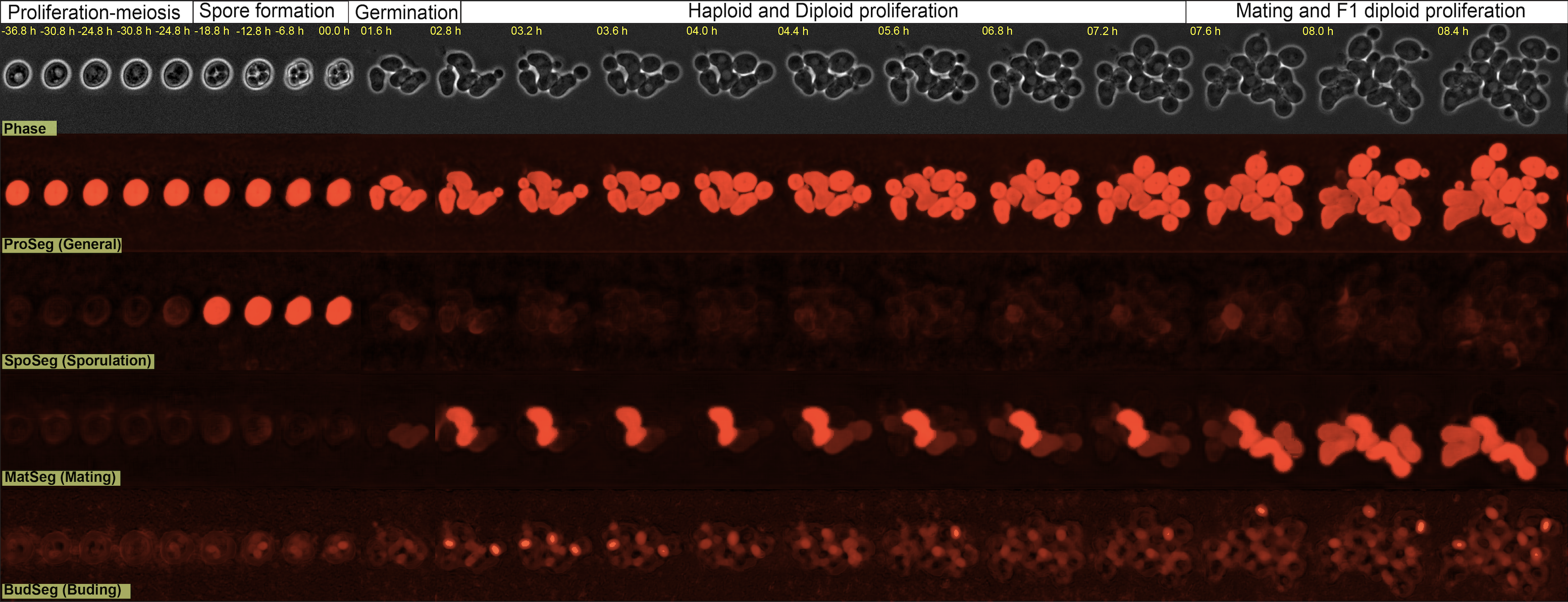
- Segment cytoplasm, vacuoles, buds, mating, and sporulating yeast cells
- Extract and plot time-series data in the GUI
This package supports Linux, Windows and Mac OS (versions later than Yosemite). GPU support is available for NVIDIA GPU's. A GPU is recommended, but not required, to run yeastvision
yeastvision is ready to go for cpu-usage as soon as it downloaded. GPU-usage requires some additional steps after download. To download:
- Install an Anaconda distribution of Python. Note you might need to use an anaconda prompt if you did not add anaconda to the path.
- Open an anaconda prompt/command prompt
- If you have an older
yeastvisionenvironment you should remove it withconda env remove -n yeastvisionbefore creating a new one. - Create a new environment with
conda create --name yeastvision python=3.10.0. - Activate this new environment by running
conda activate yeastvision - To download our package plus all dependencies, run
python -m pip install yeastvisionon Windows andpython3 -m pip install yeastvisionon Linux, Ubuntu, and Mac OS.
You should upgrade the yeastvision package periodically as it is still in development. To do so, run the following in the environment:
python -m pip install yeastvision --upgradefor Windows or
python3 -m pip install yeastvision --upgradeSee Troubleshooting: Common Problems for potential install errors.
To use your NVIDIA GPU with python, you will first need to install a NVIDIA driver for
your GPU. Once downloaded, ensure that your
GPU is detected by running nvidia-smi in the terminal.
yeastvision relies on pytorch for implementation of the deep-learning models, which we will need to configure for gpu usage. Ensure your yeastvision conda environment is active for the following commands.
First, we need to remove the CPU version of torch:
pip uninstall torch
And the cpu version of torchvision:
pip uninstall torchvision
Now install torch and torchvision for CUDA version 11.3. Ensure that your nvidia drivers are up to date for version 11.3 by running nvidia-smi and check that a version 11.3 or greater is displayed in the top right corner of the output table.
conda install pytorch==1.12.0 torchvision==0.13.0 cudatoolkit=11.3 -c pytorch
After install you can check conda list for pytorch, and its version info should have cuXX.X, not cpu.
Activate the correct conda environment. In the command prompt, run either
yeastvision -testto open the GUI with our sample 10-frame movie.yeastvisionto open a blank GUI window. Drag and drop directory containing 2D-image files into the GUI to begin segmenting, labeling, or processing
Yeastvision accepts directories of image files, loaded through drag-and-drop or the file menu. Each file in the directory should contain only a single 2D image, named with a standard image file extension.
Note: As you segment, interpolate, or process images, GUI-generated labels and images are stored in the loaded directory as .npz files. Deleting these files results in loss of the data.
Yeastvision expects directories to contain only files with valid image extensions or .npz files created in previous yeastvision sessions.
Important: Ensure that all images in the same channel have the same dimensions.
To load directories containing additional image channels and pre-generated masks, name your files properly:
For experiments containing multiple channels and pre-generated labels, our GUI loading capabilities make it easy to:
- Track pre-generated labels (a nucleus label, for example)
- extract and plot fluorescence intensities
- interpolate additional fluorescence channels.
When additional channels/masks are present in the directory, files should be named accordingly to their channel and mask type:
- Each data type in the directory should have a standard id that directly precedes the file extension (ex: _phase.tif, _channel2.png, _mask1.tif)
- Any image that acts as a label should have
_maskin the id (ex: _mask_nucleus.tif, _mask_cytoplasm.jpg). - Ensure that each id has the same number of time points: Having 5 phase images but only 4 fluorescence images will raise an error.
Here is an example of a directory with two time points, two channels, and two pre-generated labels, sorted by name:
im001_channel1.tif, im001_channel2.tif,
im001_mask1.tif, im001_mask2.tif,
im002_channel1.tif, im002_channel2.tif,
im002_mask1.tif, im002_mask2.tif
Yeastvision contains models to accurately segment yeast in all stages of their lifecycle. Simply choose one of the following models from the model dropdown and click run.
Pixel flow-based models
| Model | Segments |
|---|---|
| proSeg | proliferating cells (general cytoplasm segmentation) |
| spoSeg | sporulating cells |
| matSeg | mating cells |
| budSeg | bud-necks |
- Ensure that training data is loaded into the current experiment
- Select the model to be retrained from the mainscreen model dropdown
- Click Menu->Models->Retrain
- Ensure training data is correct and choose model suffix (default is date-time)
- Select hyperparameters (default should work for most use cases)
- Train the model, using terminal to gauge progress.
- The custom model will auto-run on the next available image in the training set, if there is not a mask already on this image.
- If you are happy with the new model, go to Menu->Models->Load Custom Models, and the model will be added to the model dropdown. Otherwise, retrain with new data
- Even though it possible to retrain using only CPU, training takes very long without a GPU
- When you are initially producing a training set, leave some blank masks towards the end of the movie so that the training procedure has room to auto-run
- The path to the new weights will be printed on the terminal.
- Ensure that the fullname of the retrained model is present in the weights filename upon trying to load it via the models menu. This ensures that GUI can associate the weights with the correct model architecture
- Optional: Interpolate images to increase resolution, generating intermediate frames that improve tracking accuracy
- Segment and track the interpolated frame. Tracking automatically generates a cell data table that includes various morphological and image properties of each cell over each frame in the movie.
- Optional: reconstruct proliferating cell lineages
- Remove interpolated frames from the mask, so that only frames present in the original movie exist in the final tracked movie
- Produce some initial plots of the data using the 'show plot window' button. This will allow you to view single cell and population averages over time.
| Command | Function |
|---|---|
| up/down | scroll through channnels |
| cntrl + up/down | Scroll through labels |
| right/left arrows | scroll through timeseries |
| cntrl + right/left | scroll through timeseries by 3 |
| O | outline Drawing |
| B | brush Drawing |
| E | eraser |
| . | increment brush size |
| , | decrecement brush size |
| Delete/Backspace | Delete Selected Cell |
| c | show current label contours |
| f | toggle probability (if present) |
| space bar | toggle mask display |
| p | show plot window |
| Problem | Solution |
|---|---|
ModuleNotFoundError: No module named 'chardet' |
run pip install chardet |
| Cannot scroll through images/masks on the display | Click on the display to bring focus back to this widget |
| Loaded images without masks but cannot draw | An existing label must be present to draw: Add a blank label with File -> Add Blank Label |
[1] Huang, Z., Zhang, T., Heng, W., Shi, B., & Zhou, S. (2022). Real-time intermediate flow estimation for video frame interpolation. In Proceedings of the European Conference on Computer Vision (ECCV).