Home
:[toc]:
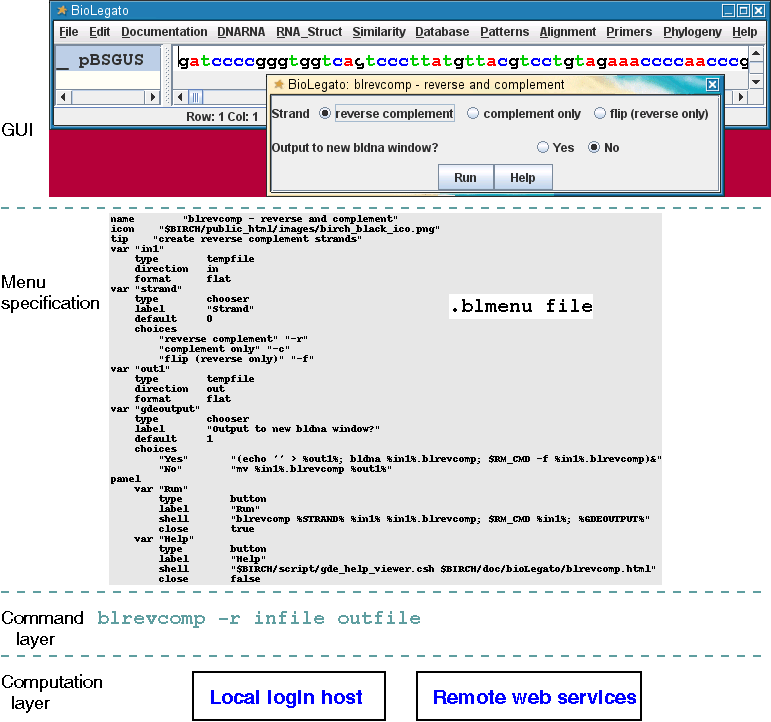
BioLegato is a generic graphical user interface (GUI), in essence, a program that runs other programs. Data are displayed and selected in a canvas. A program is chosen from dropdown menus, parameters are set in a pop up menu, and the program launched by issuing a shell command to the system. Thus, any program, from a simple shell script to a complex analytical pipeline, can be launched from BioLegato.
BioLegato is programmable in the sense that for each external program, a parameter menu, and its corresponding shell command, are specified using a simple language called PCD. PCD makes it easy even for non-programmers to specify which parameters are set, their default values or choices.
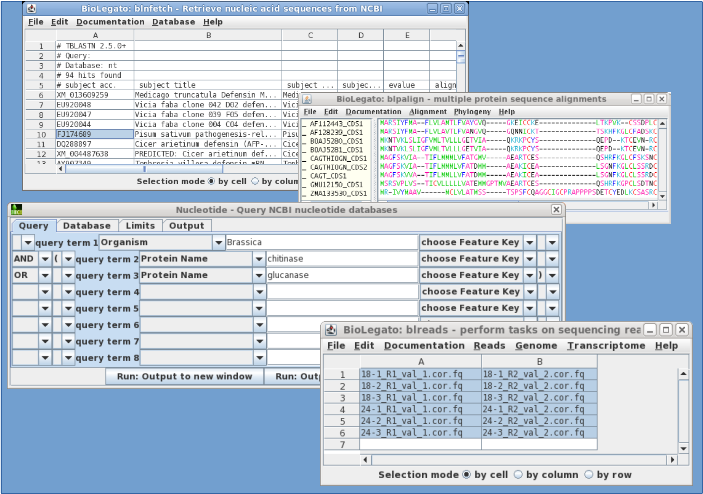
BioLegato was written as the graphical user interface for the BIRCH Bioinformatics System. BioLegato can work with many different kinds of data, including sequences, alignments, phylogenetic trees, molecular markers, and NGS sequencing reads.
BioLegato makes it easy to point and click through complex analytical pipelines, such as genome or transcriptome assembly.
For each type of data, you can create a new BioLegato application by combining a canvas appropriate to the data with external programs that work on that type of data.
BioLegato calls programs that run as external jobs. In essence, any program can be run from BioLegato. Programs can run on a local server, or as a remote web service. Output is returned to the user's desktop in the appropriate viewer.
As illustrated below, each menu in a BioLegato application is specified in a file. The file contains BioPCD code describing widgets to be included in the menu, default values, and a shell command to be executed using the parameters set in the menu.
We welcome applications from potential developers who would like to contribute to the BioLegato project. In particular, we are interested in developers who would like to adapt BioLegato to run programs in other fields in science and engineering.
Potential development goals:
- New Biolegato applications for different types of data
- New BioLegato canvases for different types of data
- New features for the PCD language
- Bug fixes
BioLegato Development Protocols
Features
Alvare GGM, Roche-Lima A, Fristensky B (2012) BioPCD - A Language for GUI Development Requiring a Minimal Skill Set Int. J. Computer Appl. 57:9-16. doi: 10.5120/9117-3273
Alvare, G., Roche-Lima, A. & Fristensky, B. BioLegato: a programmable, object-oriented graphic user interface. BMC Bioinformatics 24, 316 (2023). https://doi.org/10.1186/s12859-023-05436-4