-
Notifications
You must be signed in to change notification settings - Fork 170
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Integrating BrainDecode in MOABB benchmarking pipelines #46
Comments
|
What is status of this is it still planned? @sylvchev |
|
It is still planned. |
|
@agramfort continuing the discussion from the PR,
|
|
I cannot invest time to adapt your code to our computer cluster. We don't
work
with notebooks in the team to run batch trainings. I would need 2 scripts:
- 1 that downloads and put the files in one place
- 1 that runs the code
Readme should explain to me what versions of the packages to use
… |
|
Sure, I should be able to do that. To add the braindecode models should I make it such that they test on the existing datasets of moabb? And should I add a run for benchmarking the sleep models on the physionet sleep dataset? |
|
I was attempting running a Braindecode model in MOABB's evaluation and ran into an error while fitting the pipeline/classifier. Error - Is it something in the way I'm doing it? |
|
Ah yes, I was doing with the PyPI version, using the master version solved that. I was able to create the dataset and pass it to the classifier, but I'm getting this weird error with on_epoch_end where the version of skorch is seemingly checked by braindecode I think, I tried the dev version of skorch after I got this error with the PyPI version and get the same error with both versions. and the error comes with the version checking as this - Don't mind the printed stuff on the Colab notebook cell output above this error, that was just something for debugging and is not relevant to the error. |
|
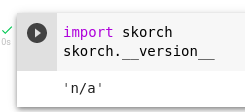
So it seems skorch returns version 'n/a' for some reason the way it was installed in your linked colab: Seems this happens here: Unclear to me why it happens precisely, we could fix on our side by changing: braindecode/braindecode/training/scoring.py Line 358 in dba19a7
to if (skorch.__version__ != 'n/a') and (not check_version('skorch', min_version='0.10.1')):(basically assuming n/a means we are up to date with skorch, maybe good to put a comment there) |
|
@robintibor I checked again today with the 0.11.0 PyPI version of skorch that was pushed 2 days back and the error doesn't occur anymore. |
|
well @Div12345 yes I think this can only happen with the master version of skorch, but maybe we still want to be compatible in any case? |
|
@robintibor I'll make the check for the other versions but is it safe to assume that NaN implies that skorch is up to date? Couldn't it just mean that the version check is just failing somewhere and is uncertain? In which case, at least we could make a warning that the version is uncertain so that if some error occurs people can understand the probable cause more easily? |
|
Hello guys! I am trying to close open issues in braindecode, and I spent some time on this issue. I think I have found the solution to this issue. Based on the @Div12345 code, I made a minimal viable code that integrates Moabb and Braindecode. It still needs to be well optimized! The code is below. import os.path as osp
import matplotlib.pyplot as plt
import mne
import seaborn as sns
import torch
from braindecode import EEGClassifier
from braindecode.datasets import create_from_X_y
from braindecode.models import ShallowFBCSPNet
from braindecode.util import set_random_seeds
from moabb.datasets import BNCI2014001
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import LeftRightImagery
from moabb.utils import set_download_dir
from numpy import unique
from sklearn.base import BaseEstimator, ClassifierMixin, TransformerMixin
from sklearn.pipeline import Pipeline
from skorch.callbacks import LRScheduler
set_download_dir(osp.join(osp.expanduser("~"), "mne_data"))
cuda = (
torch.cuda.is_available()
) # check if GPU is available, if True chooses to use it
device = "cuda" if cuda else "cpu"
if cuda:
torch.backends.cudnn.benchmark = True
seed = 20200220 # random seed to make results reproducible
# Set random seed to be able to reproduce results
set_random_seeds(seed=seed, cuda=cuda)
n_classes = 2
# hard-coded for now
n_chans = 22
input_window_samples = 1001
model = ShallowFBCSPNet(
n_chans,
n_classes,
input_window_samples=input_window_samples,
final_conv_length="auto",
)
# Send model to GPU
if cuda:
model.cuda()
# These values we found good for shallow network:
lr = 0.0625 * 0.01
weight_decay = 0
batch_size = 64
n_epochs = 4
clf = EEGClassifier(
model,
criterion=torch.nn.NLLLoss,
optimizer=torch.optim.AdamW,
train_split=None, # using valid_set for validation
optimizer__lr=lr,
optimizer__weight_decay=weight_decay,
batch_size=batch_size,
callbacks=[
"accuracy",
("lr_scheduler", LRScheduler("CosineAnnealingLR", T_max=n_epochs - 1)),
],
device=device,
)
class Transformer(BaseEstimator, TransformerMixin):
def __init__(self, kw_args=None):
self.kw_args = kw_args
def fit(self, X, y=None):
self.y = y
return self
def transform(self, X, y=None):
dataset = create_from_X_y(
X.get_data(),
y=self.y,
window_size_samples=X.get_data().shape[2],
window_stride_samples=X.get_data().shape[2],
drop_last_window=False,
sfreq=X.info["sfreq"],
)
return dataset
def __sklearn_is_fitted__(self):
"""Return True since Transfomer is stateless."""
return True
class ClassifierModel(BaseEstimator, ClassifierMixin):
def __init__(self, clf, kw_args=None):
self.clf = clf
self.classes_ = None
self.kw_args = kw_args
def fit(self, X, y=None):
self.clf.fit(X, y=y, **self.kw_args)
self.classes_ = unique(y)
return self.clf
def predict(self, X):
return self.clf.predict(X)
def predict_proba(self, X):
return self.clf.predict_proba(X)
create_dataset = Transformer()
fit_params = {"epochs": 10}
brain_clf = ClassifierModel(clf, fit_params)
# from functools import partial
# clf.fit = partial(clf.fit, epochs=10)
pipe = Pipeline([("Braindecode_dataset", create_dataset), ("Net", brain_clf)])
print(pipe)
pipes = {}
pipes["ShallowFBCSPNet"] = pipe
mne.set_log_level(False)
# Define Evaluation
paradigm = LeftRightImagery()
# Because this is being auto-generated we only use 2 subjects
dataset = BNCI2014001()
dataset.subject_list = dataset.subject_list[:2]
datasets = [dataset]
overwrite = True # set to True if we want to overwrite cached results
evaluation = WithinSessionEvaluation(
paradigm=paradigm,
datasets=datasets,
suffix="braindecode_example",
overwrite=overwrite,
return_epochs=True,
)
results = evaluation.process(pipes)
print(results.head())
##############################################################################
# Plot Results
# ----------------
#
# Here we plot the results. We the first plot is a pointplot with the average
# performance of each pipeline across session and subjects.
# The second plot is a paired scatter plot. Each point representing the score
# of a single session. An algorithm will outperforms another is most of the
# points are in its quadrant.
fig, axes = plt.subplots(1, 2, figsize=[8, 4], sharey=True)
sns.stripplot(
data=results,
y="score",
x="pipeline",
ax=axes[0],
jitter=True,
alpha=0.5,
zorder=1,
palette="Set1",
)
sns.pointplot(
data=results, y="score", x="pipeline", ax=axes[0], zorder=1, palette="Set1"
)
axes[0].set_ylabel("ROC AUC")
axes[0].set_ylim(0.5, 1)
# paired plot
paired = results.pivot_table(
values="score", columns="pipeline", index=["subject", "session"]
)
paired = paired.reset_index()
axes[1].plot([0, 1], [0, 1], ls="--", c="k")
axes[1].set_xlim(0.5, 1)
plt.show()How do you want to proceed, @sylvchev? |
|
Thanks for this nice example. I'm working with @pangolinMagique on this integration. He just found a way to integrate braindecode using only epochs from MOABB, as you have. One central question is do MOABB need to provide access to raw signal or is epochs enough to have good performance? |
|
feel free to reopen if you feel we should discuss more.
… Message ID: ***@***.***>
|
|
Hello @sylvchev and @agramfort, I feel this is a parameter search. Currently, I am thinking about how to coordinate the team to code this integration (@agramfort, @bruAristimunha, @sylvchev and @pangolinMagique). We can conduct a small study, maybe reproducing the robin results with moabb and braindecode. I was wondering, shall we continue this topic via e-mail or braindecode chat instead of here? I think that coordinating it escapes the context of the issue. |
|
I prefer if conversations are in the open but we can maybe zoom at some
point to simplify things
… Message ID: ***@***.***>
|
|
No problem, we'll solve it here then. |
|
Yes, good idea, it will be more efficient |
|
We have now the possibility to expose raw data to train braindecode models in MOABB (see NeuroTechX/moabb#302) |
|
@bruAristimunha I think that @sylvchev is much more competent to review here. I trust your judgement. Feel free to merge if happy. @sylvchev you have the green buttons on both repos :) |
|
Thanks @agramfort |
Using the scikit-learn API, BrainDecode could be used as a regular scikit-learn pipeline and benchmarked against others classical BCI pipelines.
The text was updated successfully, but these errors were encountered: