-
Notifications
You must be signed in to change notification settings - Fork 22
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
The function detect_heartbeats is not handling the absence of signal correctly #87
Comments
|
Yes, this is a property of the Pan-Tompkins detector, which expects a heartbeat after a certain amount of time. I agree we should have some kind of (optional) preprocessing, which detects e.g. flat segments (which is relatively easy) and extremely noisy segments (which is probably hard). In such segments, no heartbeats should be detected. The thresholds of the detector should probably remain constant until the next good data segment – but this is also open for discussion. I think one way to detect flat segments could be to only perform a certain number of threshold decreases. I think currently we do it 16x, and then we just take the peak that's in that signal. We could instead say that if there is no clear peak after e.g. 8x lowering the thresholds, we skip it. But this wouldn't be the classic Pan-Tompkins anymore, so I'd prefer a separate preprocessing step. How do other packages solve this issue? |
|
Yes, I think the classic Pan-Tompkins version implemented here should at least be fixed so that it does not run forever with signals like the one I shared, so skipping after a given number of thresholds could maybe solve this. One solution could be to have an optional |
|
I agree something should be done to address these situations. I need to think a bit more about what the best solution would be. In addition, I do not have a lot of time to work on this at the moment, but if you would like to give it a shot please go ahead! I'm happy to discuss any details and questions of course, I just don't have time to actually implement something in the near future. |
|
Hi, Just adding my two cents here. From #88, I think that this issue is only true when the beginning (not the end) of the data is flat. An easy way to check for the first non-flat sample in the ECG data is: idx_nonflat = np.nonzero(np.diff(ecg))[0]
if not len(idx_nonflat):
# The entire signal is flat. Raise an error.
else:
# Take the first non-flat index
idx_nonflat[0]Not sure where we go from here: should we raise a warning/error or just continue with the calculation using the truncated array ( Thanks, |
|
What about flat segments in the middle of the data? I always thought that the detector forcibly finds a heartbeat after a certain amount of time after the previous detection, no matter what the signal looks like. Is this not the case? Can you maybe show how it performs in the presence of flat segments at the beginning, in the middle, and at the end of the data? I think it would be important to know what the detector does before deciding how to proceed. |
import sleepecg
import numpy as np
from scipy.misc import electrocardiogram
ecg = electrocardiogram() # 5 min of ECG data
sf = 360
y_true = sleepecg.detect_heartbeats(ecg, sf)
y_true.shape # 478 true peaks
# Flat data at the end: 478 peaks detected. PASS.
ecg_pad_after = np.pad(ecg, pad_width=(0, ecg.size), mode="edge")
pks = sleepecg.detect_heartbeats(ecg_pad_after, sf)
print(f"{len(pks)} detected peaks")
# Flat data at the beginning: 1943 peaks detected. FAIL.
ecg_pad_before = np.pad(ecg, pad_width=(ecg.size, 0), mode="edge")
pks = sleepecg.detect_heartbeats(ecg_pad_before, sf)
print(f"{len(pks)} detected peaks")
# Flat data in the middle: 926 detected peaks.
# Here we expect 2 * 478 = 956 peaks since we have duplicated the data.
ecg_pad_middle = np.hstack((ecg_pad_after, ecg))
pks = sleepecg.detect_heartbeats(ecg_pad_middle, sf)
print(f"{len(pks)} detected peaks") |
|
Detection critically relies on the two thresholds. If these are bad, it takes a long time for the detector to recover. That's why padding afterwards works, padding in the middle works OK, and padding at the beginning doesn't work but eventually yields correct peak towards the end. So "flat data in the middle" should not get a ❌, but rather a ≈ (the expected number of peaks is 478 × 2 = 956). import matplotlib.pyplot as plt
import numpy as np
from scipy.misc import electrocardiogram
import sleepecg
ecg = electrocardiogram() # 5 min of ECG data
sf = 360
y_true = sleepecg.detect_heartbeats(ecg, sf)
y_true.shape # 478 true peaks
fig, axes = plt.subplots(3, 1, figsize=(16, 8))
# Flat data at the end: 478 peaks detected. PASS.
ecg_pad_after = np.pad(ecg, pad_width=(0, ecg.size), mode="edge")
t_pad_after = np.arange(0, len(ecg_pad_after) / sf, 1 / sf)
pks_after = sleepecg.detect_heartbeats(ecg_pad_after, sf)
axes[0].plot(t_pad_after, ecg_pad_after)
axes[0].plot(pks_after / sf, ecg_pad_after[pks_after], "r.")
print(f"{len(pks_after)} detected peaks")
# Flat data at the beginning: 1943 peaks detected. FAIL.
ecg_pad_before = np.pad(ecg, pad_width=(ecg.size, 0), mode="edge")
t_pad_before = np.arange(0, len(ecg_pad_before) / sf, 1 / sf)
pks_before = sleepecg.detect_heartbeats(ecg_pad_before, sf)
axes[1].plot(t_pad_before, ecg_pad_before)
axes[1].plot(pks_before / sf, ecg_pad_before[pks_before], "r.")
print(f"{len(pks_before)} detected peaks")
# Flat data in the middle: 926 detected peaks. FAIL.
ecg_pad_middle = np.hstack((ecg_pad_after, ecg))
t_pad_middle = np.arange(0, len(ecg_pad_middle) / sf, 1 / sf)
pks_middle = sleepecg.detect_heartbeats(ecg_pad_middle, sf)
axes[2].plot(t_pad_middle, ecg_pad_middle)
axes[2].plot(pks_middle / sf, ecg_pad_middle[pks_middle], "r.")
print(f"{len(pks_middle)} detected peaks") |
|
Oh right, thanks for catching my mistake with the flat data in the middle. Of course we're expecting twice the number of peaks 🤦♂️ |
And these two thresholds are calculated somewhere in the beginning of the data? So do you think my above suggestion of starting the detector at the first non-flat index would be good enough? |
|
Yes. It would be worth a shot. |
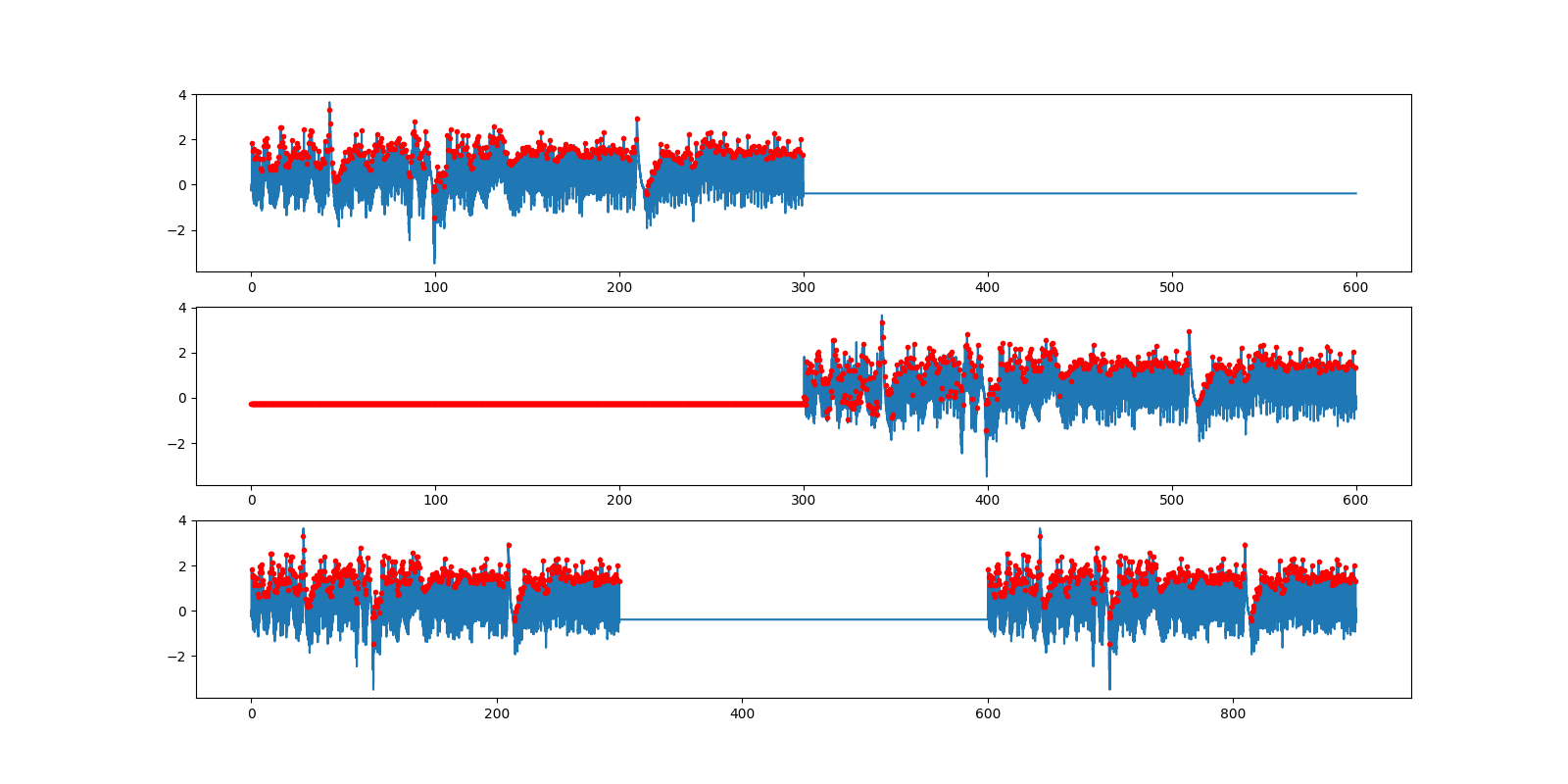
The function
detect_heartbeats()can be easily confused when presented with time series containing a flat signal e.g. at the beginning of the recording.The problem is that the flat signal at the beginning or at the end of the time series is not discarded automatically. Of course, better performance is achieved by providing better quality data, but I think this is something that should be automatically handled by the function: no peak detection on the portion of the signal consisting of consecutive constant values over at least n samples, at the beginning or at the end (ideally also if this happened during the recording).
In addition to that, the erroneous detection of (lot of) peaks in these segments appears to considerably slow down the function itself, the following time series (~1h ECG recording starting with ~45 s of flat signal) can for example completely crash the function that will hang forever (or at least for a very long time).
ecg.zip
The text was updated successfully, but these errors were encountered: