New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
misassembly introduced in hifiasm v0.5 #10
Comments
|
Thanks so much for point that. It seems that the mis-assemblies are caused by the our new purge_dup. I will expose its parameters to users today, and let you know when it is available. |
|
v0.5 fixes one misassembly on our data, but generally hifiasm does produce large misassemblies occasionally. @chhylp123 will expose some purge_dups parameters, which may help. We are also thinking about the possibility to integrate some bogart heuristics in future. |
|
I have exposed three purge_dups parameters: '-l', '-s', '-O'. I guess it may help for fixing assemblies. Please use the latest commit with version 0.5-dirty-r247 (hash: 7f6725e). Looking forward to your results @HenrivdGeest . |
|
Thanks, I will start a sweep asap. Is their a way to re-use the error corrected reads? With some parameter testing, it can be faster to run that step once. |
|
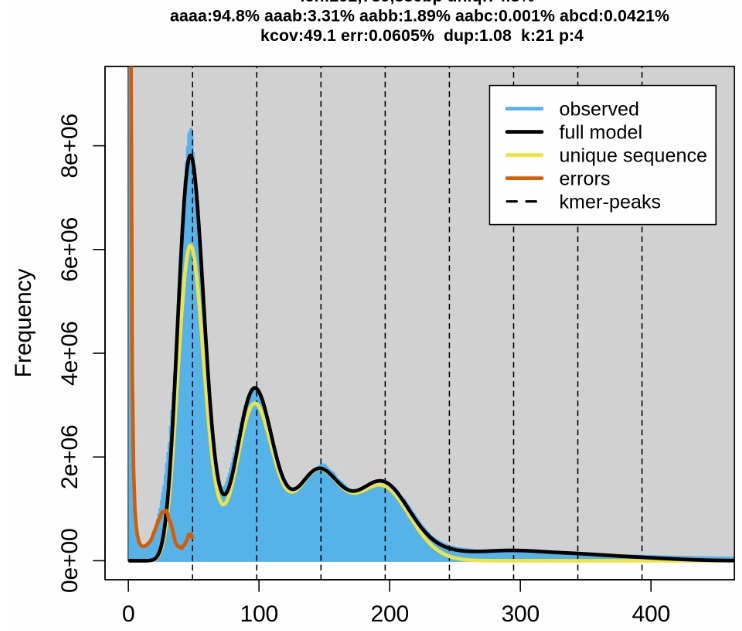
If you specify the same "-o prefix" option, hifiasm will reuse "prefix.*.bin" files and skip error correction and overlapping. Note that older assembly graphs will be overwritten if you do this. By the way, what is the heterozygosity of your genome? v0.5 outputs multiple k-mer histograms in stderr. Is it possible to show us the first histogram? Thanks. |
|
Thanks a lot! Is this public data? We have been mostly using animal data for testing. The heterozygosity is much lower in comparison to some plant genomes. While redwood heterozygosity is high (higher than your genome) and its data is public, it takes several days to assemble, making testing very difficult. If your data is public or can be shared with us privately, it would be ideal for development purposes. Some hifiasm parameters are tuned for low heterozygosity, we may have a lot of room for improvement. Also, what is the preferred output for your genome? Is it one primary assembly representing one haplotype? |
|
I made 18 assemblies, all combinations of -l (0,1,2) -s (0,75,0,90) and -O (1,5,10). But all of them show exactly 2 misassemblies. I measured my version 0.3 the same, and that one has none. The n50's of 0.3 and the "v0.5 -l0,-s0.75 -l1" version are almost equal: |
|
I see. Thanks a lot. Let me check what's the difference between v0.3 and v0.5... |
|
I think you can have try to further increase -s and -O. I guess even '-s 0.99' is fine for hifiasm. Actually '-s 1' doesn't mean hifiasm only purges exactly same haplotigs, it still allows differences. |
|
I tried increasing -s to 0.999 and -O to 75, but that did not made any difference in the output regarding the misassembly. I tried running v05 on the v03 bin files, but for v03 I only have a *reverse.bin, and that alone doesn't seem to work, I think it starts assembling from scratch. |
|
Thanks a lot, I will expose another option to users. I believe that can avoid these two mis-assemblies. |
|
BY any change any luck on these new options? |
|
Oh, I'm so sorry I forget that : ( . I will expose it this day. |
|
Please wait me one more day, I will fix it soon. Thanks a lot : ) |
|
I have exposed '-u' to disable post-joining (0.7-dirty-r256). Hope it is helpful. I'm so sorry for the deay : (. |


I am following hifiasm for our plant genome assembly, and noticed that with my code update from hifiasm v03 to v05 the assembly n50 increased but in my genome 2 appearant misassemblies got my attention.


I highlight one in the images below. We have a tetraploid plant of ~ 400MB haploid size, and we have >100x hifi coverage of the haploid genome, meaning ~25x per haplotype.
I already assembled this genome with hiCANU, and noticed also there that with changing the bogart (assemble) parameters too much, I easily introduce misassemblies. As far as I know, I can't alter the settings for hifiasm.
In this image you see the alignment of the contigs to a related public reference, with the assembly error:
If I look at the same positions (zoom in) but for the unitigs; I see that the unitigs do not contain this error:
Are their any ways to make the contigging more stringent? (version 0.3 did not have this error yet)
I have to say that I am not 100% sure this is not a true biological case, I am confindent that this is an assembly error, also from what I've seen with hiCanu. If you need more info, let me know
The text was updated successfully, but these errors were encountered: