-
Notifications
You must be signed in to change notification settings - Fork 97
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
BA.5.5 sublineage with S:N450D (201 seq. on Usher as of 08.08.2022) #873
Comments
|
Great proposal - seems to be a mutation of interest indeed. I'd do growth advantage limited to US for now, since otherwise you get too much influence of upload speeds from different countries. Also, it's super helpful to put date of sequence count in the title, at least I find that incredibly useful when scanning issues. |
|
Hi @agamedilab the route from BA.5.5 root is C5512T then Orf8:V62L then S:450D |
|
@FedeGueli many thanks for spotting this! You are right, the S:N450D clade is apparently a sublcade of the G28077T (ORF8:V62L) clade. |
|
Thanks @agamedilab As outlined here, new Pango lineages need to be associated with both an evolutionary event and an epidemiological event. These epidemiological events can include movement of the virus into a new geographical region, rapid and sustained growth in frequency compared to other co-circulating lineages, a jump into a novel host species and acquisition of a set of mutations of particular biological interest. Pango lineages therefore track more than just amino acid mutations. There doesn't seem to be a clear epidemiological event associated with this clade as there isn't a clear change in geographical location and the proportion of BA.5.5 sequences in this clade in the USA appears to have plateaued currently: We therefore haven't designated this at this point |
|
Good morning @chrisruis, dont you think it is a bit early to shut this down? As @corneliusroemer noted the lineage carries with S:N450D a potential "mutation of interest". Also the proposal is only a week old and the clade grew by 33% during the last 7 days (from 105 to 140 genomes on Usher). Likewise the sublineage has still a growth advantage over BA.5.5 also in the US:
Therefore i dont see the urgency of shutting this down (or the harm of letting it run a little longer). Anyway i will continue to monitor the lineage and provide updates if need be. |
|
S:450D seems one of the few spike mutations to add some apparent advantage to BA.5 beyond 346X 444X and 460X |
|
Please feel free to reopen if there's an epidemiological event associated with this in the future |
|
I do agree that this sublineage seems to have a growth advantage over BA.5.5* but with 140 sequences this could still be an artefact so I think we should monitor and designate if the growth advantage becomes more stably positive. Right now, most growth seems to be in Arizona in the US. US sequences loads so we should know in a week or two. I'll reopen and tag with |
|
201 seq. on Usher as of 08.08.2022.
Growth advantage vs. BA.5.5 global 6 months: |
Added new lineage BA.5.5.1 from #873 with 93 new sequence designations, and 0 updated designations
|
Thanks for submitting. We've added lineage BA.5.5.1 with 93 newly designated sequences. Defining mutation A22910G (S:N450D). |








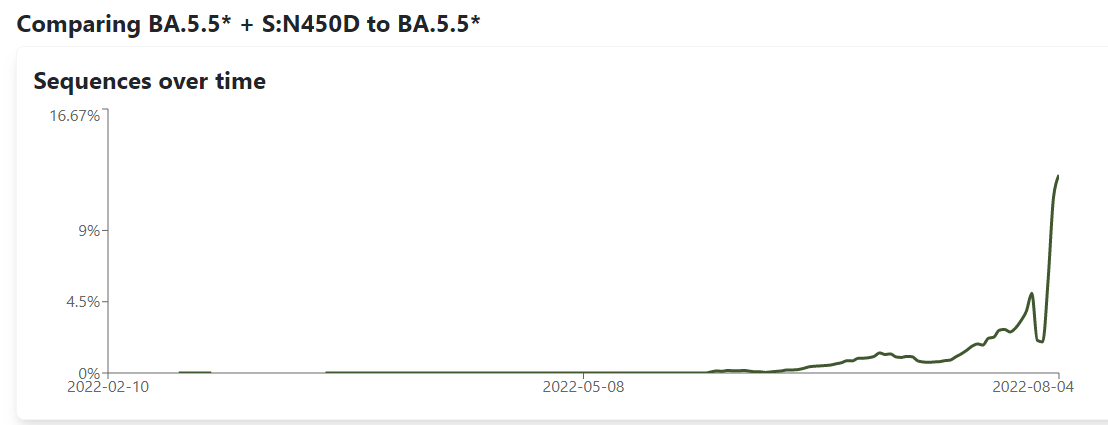



Proposal for a sublineage of BA.5.5
Earliest sequence: 04.06.2022 (USA)
Countries detected: mostly USA, UK, Israel, Germany, Mexico, Austria
Defining mutation:
A22910G = S:N450D
This variant is defined by S:N450D and was first detected in USA. Now it is present in five other countries across two continents (105 seq on Usher as of 25.07.22).
Apparently there is a lot of convergent evolution taking place with regards to S:N450D as this mutation is concurrently arising in multiple "distant" BA.2 (#851, #772, #812, #814) and BA.5 (#843) lineages.
https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_f847_e6e070.json?branchLabel=aa%20mutations&c=gt-S_450&label=nuc%20mutations:C241T,T670G,C2790T,C3037T,G4184A,C4321T,C5512T,C9344T,A9424G,C9534T,C10029T,C10198T,G10447A,C10449A,G12160A,C12880T,C14408T,C15714T,C17410T,A18163G,C19955T,A20055G,C21618T,C21789T,T22200G,G22578A,C22674T,T22679C,C22686T,A22688G,G22775A,A22786C,G22813T,T22882G,T22917G,G22992A,C22995A,A23013C,T23018G,A23055G,A23063T,T23075C,A23403G,C23525T,T23599G,C23604A,C23854A,G23948T,A24424T,T24469A,C25000T,C25584T,C26060T,C26270T,G26529A,C26577G,G26709A,C27807T,C27889T,G28077T,A28271T,C28311T,G28881A,G28882A,G28883C,A29510C
https://cov-spectrum.org/explore/World/AllSamples/Past3M/variants?aaMutations=S%3AN450D&pangoLineage=BA.5.5*&aaMutations1=S%3AN450D&pangoLineage1=BA.5.5*&
Growth advantage vs. global BA.5 3 months:
https://cov-spectrum.org/explore/World/AllSamples/Past3M/variants?pangoLineage=BA.5*&aaMutations1=S%3AN450D&pangoLineage1=BA.5.5*&analysisMode=CompareToBaseline&
EPI_ISL_13506373, EPI_ISL_13652736, EPI_ISL_13698975, EPI_ISL_13699132, EPI_ISL_13699147, EPI_ISL_13699160, EPI_ISL_13701629, EPI_ISL_13729798, EPI_ISL_13730206, EPI_ISL_13730999, EPI_ISL_13731890, EPI_ISL_13732441, EPI_ISL_13732447, EPI_ISL_13732902, EPI_ISL_13746051, EPI_ISL_13747745, EPI_ISL_13747752, EPI_ISL_13747764, EPI_ISL_13747772, EPI_ISL_13749827, EPI_ISL_13774413, EPI_ISL_13813349, EPI_ISL_13821038, EPI_ISL_13831141, EPI_ISL_13834766, EPI_ISL_13887981, EPI_ISL_13888394, EPI_ISL_13889785, EPI_ISL_13895577, EPI_ISL_13895607, EPI_ISL_13895650, EPI_ISL_13895730, EPI_ISL_13895835, EPI_ISL_13897422, EPI_ISL_13915374, EPI_ISL_13959842, EPI_ISL_13963207, EPI_ISL_13963746, EPI_ISL_14006093, EPI_ISL_14010254, EPI_ISL_14019464
ON765849.1, ON824471.1, ON867880.1, ON917879.1, ON918247.1, ON918423.1, ON918425.1, ON939869.1, ON942132.1, ON948098.1, ON948427.1, ON949452.1, ON949696.1, ON957545.1, ON966155.1, ON968270.1, ON977742.1, ON977757.1, ON978015.1, ON978017.1, ON990963.1, ON990973.1, ON991270.1, ON999612.1, ON999916.1, OP000493.1, OP009551.1, OP010133.1, OP013632.1, OP013772.1, OP014332.1, OP015030.1, OP015278.1, OP015594.1, OP016762.1, OP018010.1, OP026371.1, OP026634.1, OP026670.1, OP027373.1, OP027553.1, OP027716.1, OP027858.1, OP034538.1, OX157368.1, OX189639.1, OX221847.1, OX221897.1, OX223919.1, OX231270.1, OX231424.1, OX231442.1, OX231816.1, OX242786.1
The text was updated successfully, but these errors were encountered: