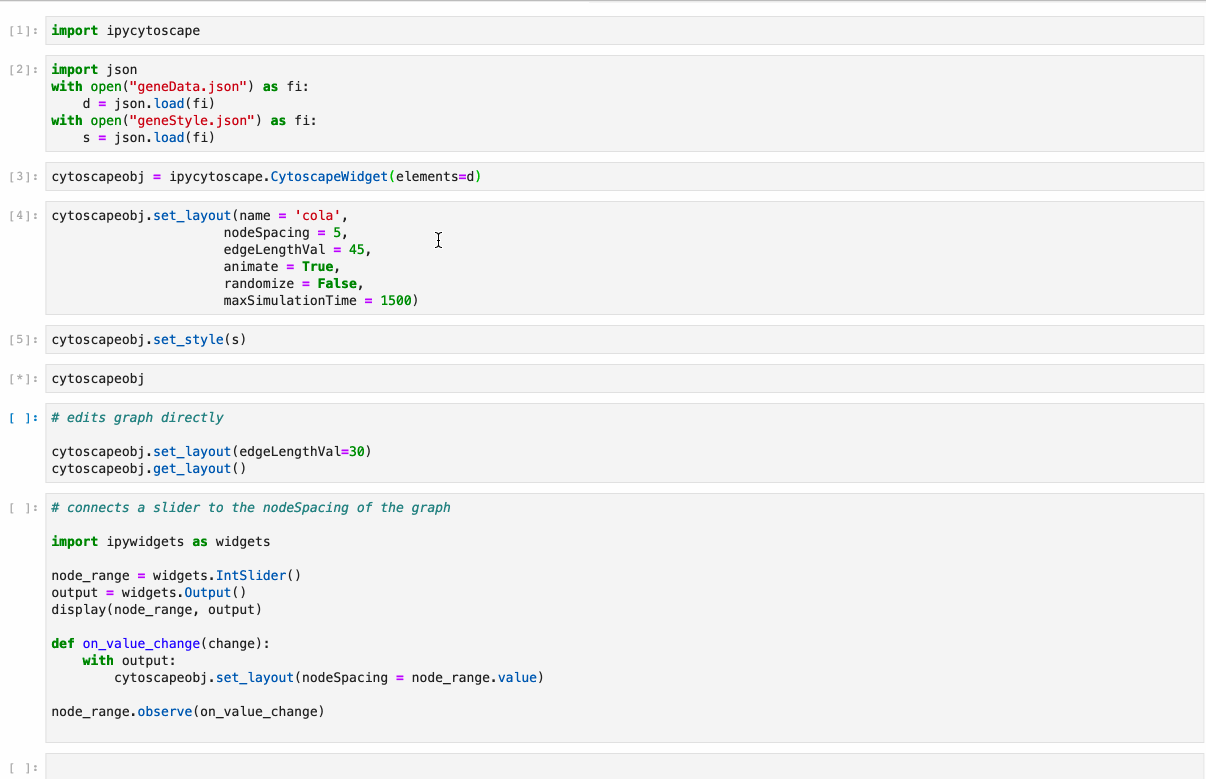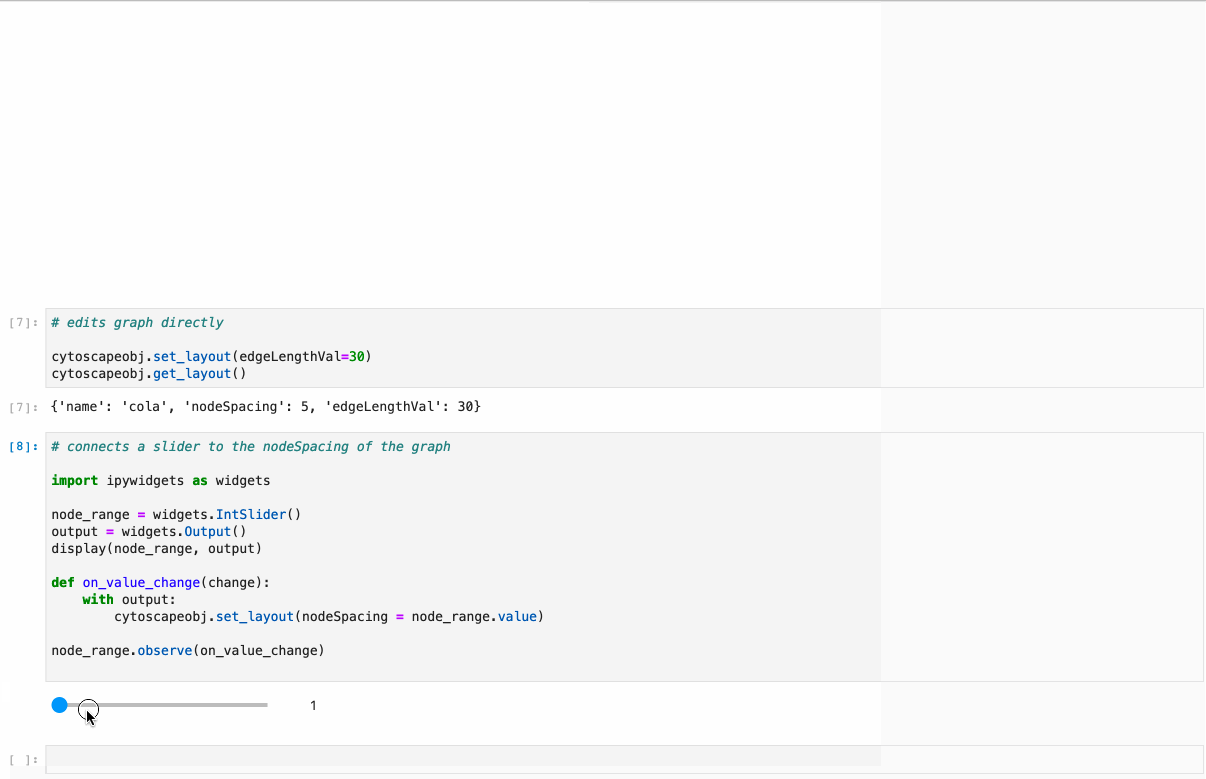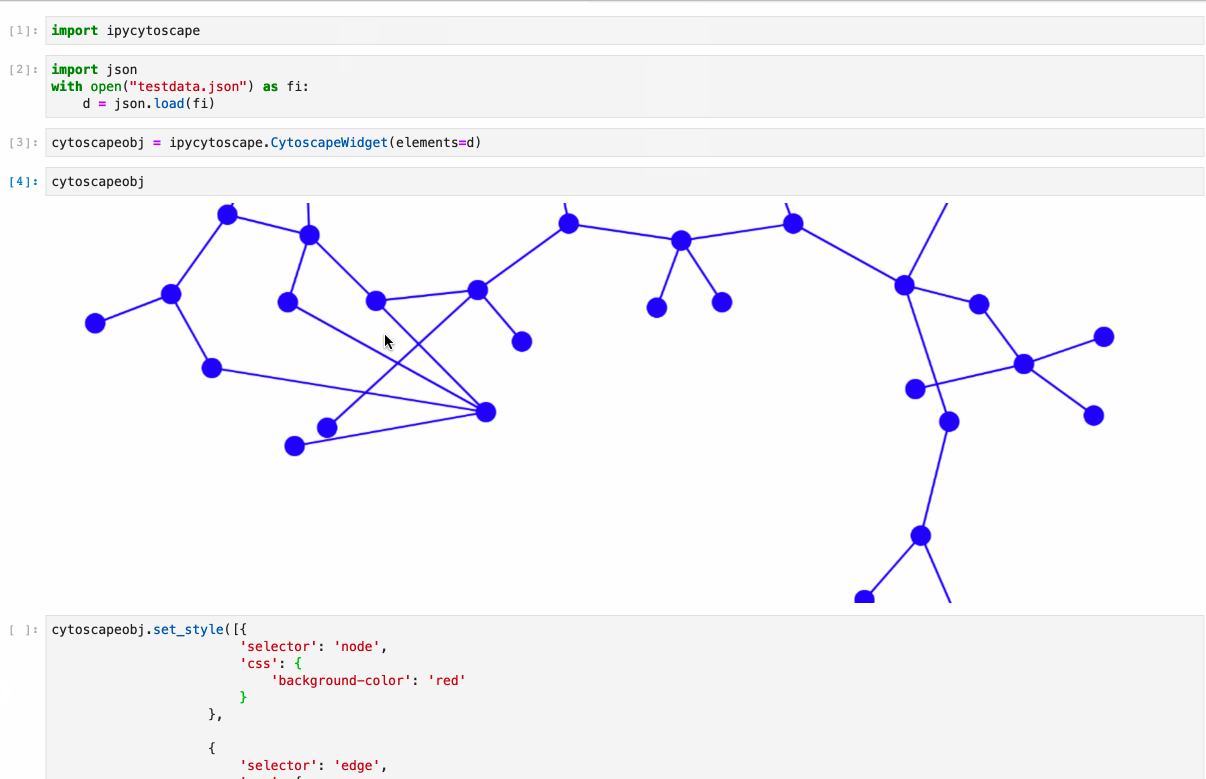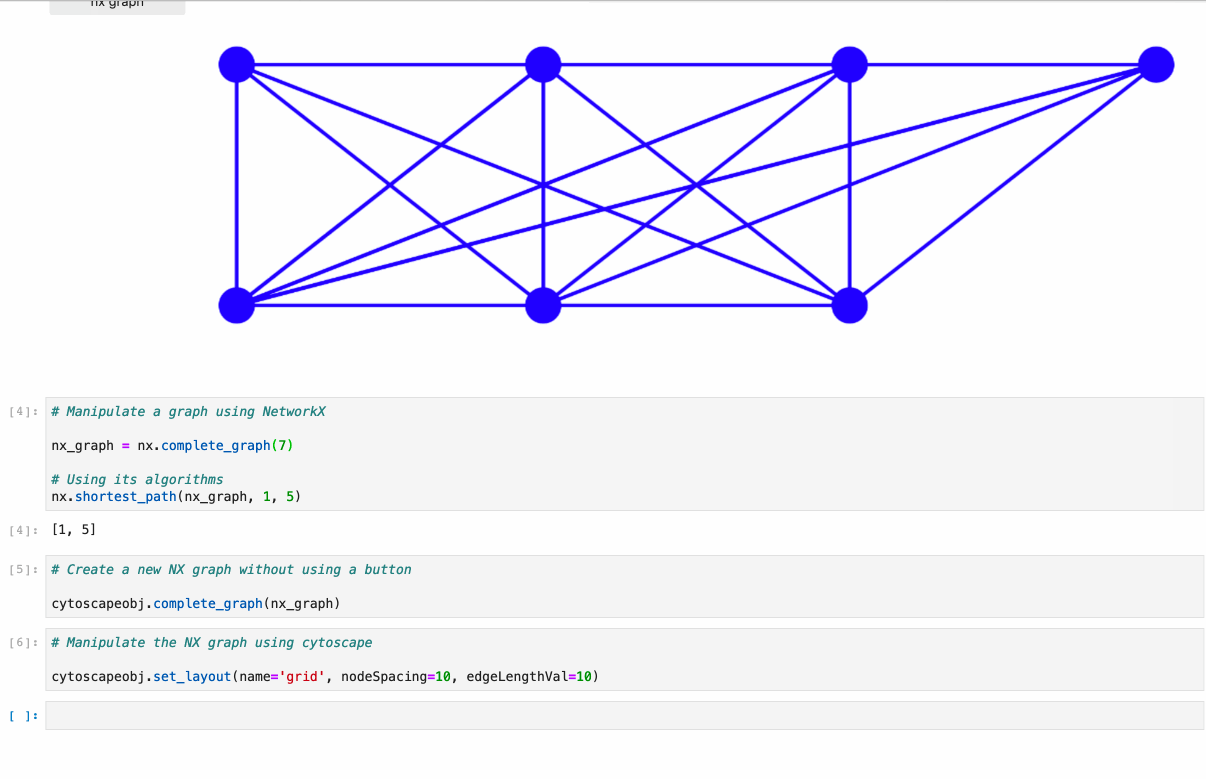
Python implementation of the graph visualization tool Cytoscape.
Offers full support to NetworkX lib. Just follow the example under /examples/Test NetworkX methods.ipynb.
With conda: (recommended)
conda install -c conda-forge ipycytoscape
With pip:
pip install ipycytoscapeOr if you use jupyterlab:
pip install ipycytoscape
jupyter labextension install @jupyter-widgets/jupyterlab-manager
jupyter labextension install jupyter-cytoscape@0.1.2If you are using Jupyter Notebook 5.2 or earlier, you may also need to enable the nbextension:
jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscapeFor a development installation: (requires npm)
$ git clone https://github.com/QuantStack/ipycytoscape.git
$ cd ipytree
$ pip install -e .
$ jupyter nbextension install --py --symlink --sys-prefix ipycytoscape
$ jupyter nbextension enable --py --sys-prefix ipycytoscape
$ jupyter labextension install @jupyter-widgets/jupyterlab-manager
$ jupyter labextension install js
We use a shared copyright model that enables all contributors to maintain the copyright on their contributions.
This software is licensed under the BSD-3-Clause license. See the LICENSE file for details.